Nationwide WastewaterSCAN February 2026 Update
Hi Everyone,
This newsletter contains updates on the WastewaterSCAN program with wastewater samples analyzed by the lab through February 27, 2026. Below you will find information on the program in terms of participating partners, review of targets being measured, as well as current wastewater categories (low, medium, high) of the infectious disease targets.
Best,
WastewaterSCAN Team
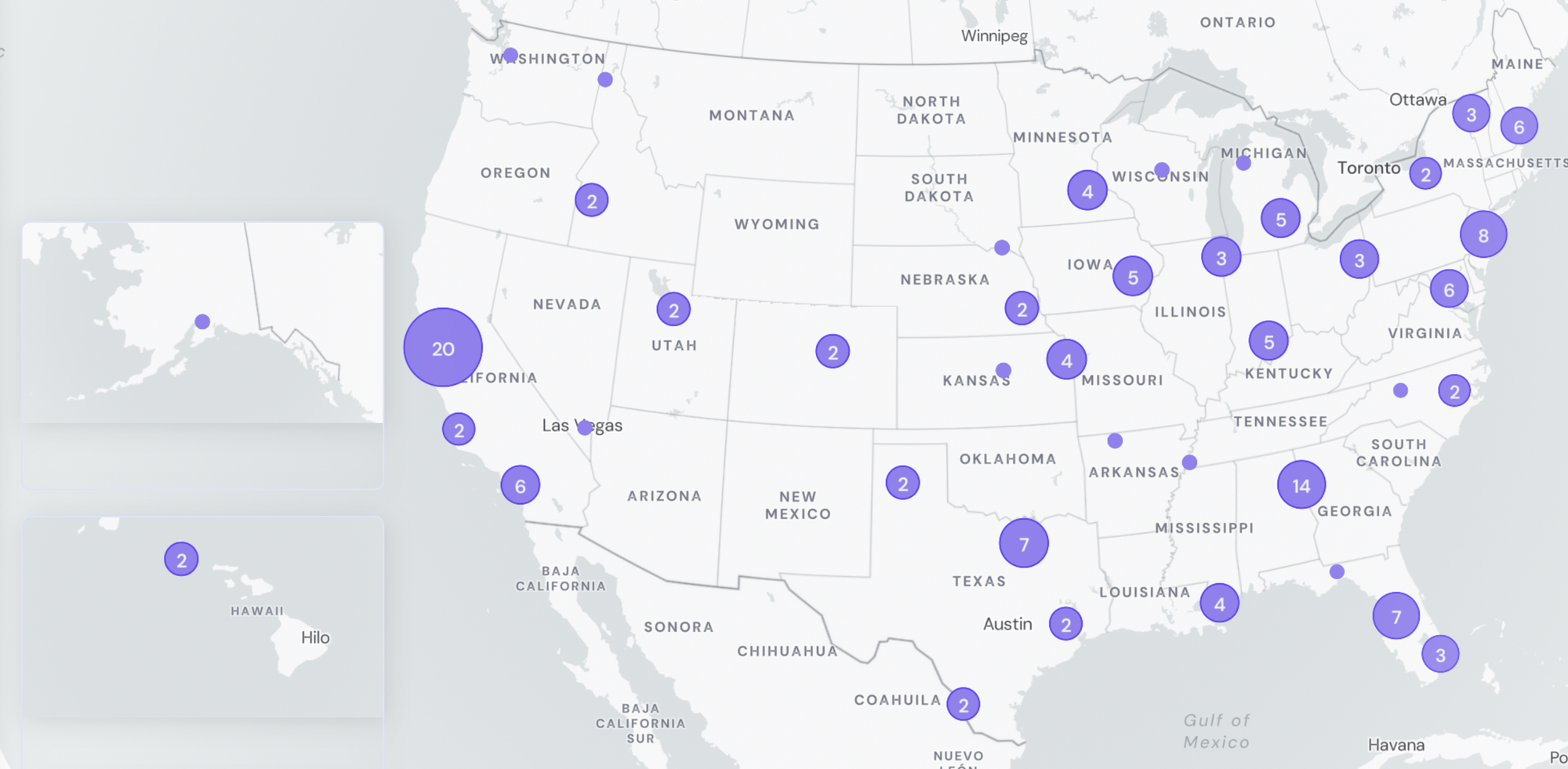
Participating Plants
147 WWTPs plants across 40 states
WWSCAN Participant Stats:
- 147 plants sampling
- >37 million people covered through sampling efforts
- 40 states
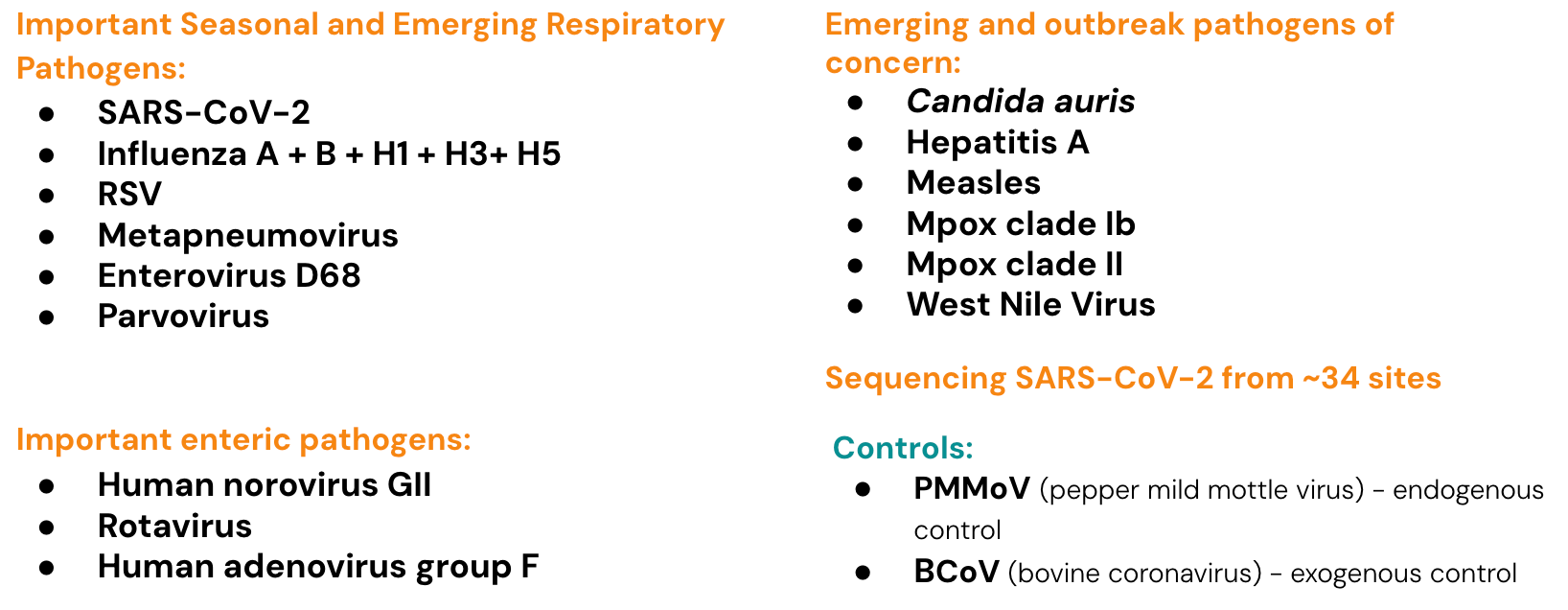
Infectious Disease Target Review
Currently monitoring a suite of Respiratory, Gastrointestinal and Outbreak Pathogens of Concern
The methods for our assays are in the public domain and links for these are provided at the end of the newsletter. West Nile Virus was added to the pathogen panel WWSCAN monitors for in mid-October 2025. As a reminder, in mid-May 2025 WWSCAN resumed measurements of Rotavirus and Adenovirus group F, and added Measles (wild-type) and Parvovirus to the list of pathogens monitored.
Program Publications
Recent publications, factsheets, FAQs, and more!
WWSCAN Published Body of Work document:
- We are excited to share a document that provides an overview of the program’s peer-reviewed scientific literature, organized by topics and themes. Click here to access the WastewaterSCAN published body of work.
WWSCAN FAQs:
- Frequently Asked Questions on Measurements of Candida auris (C. auris) in Wastewater Solids
- Frequently Asked Questions on WWSCAN Measurements of H5 Marker in Wastewater Solids
- Frequently Asked Questions on WWSCAN Measurements of Hepatitis A Virus (HAV) RNA in Wastewater Solids
- Frequently Asked Questions on WWSCAN Measurements of Measles RNA in Wastewater Solids
- Frequently Asked Questions on WWSCAN Measurements of Mpox clade Ib in Wastewater Solids
- Frequently Asked Questions on WWSCAN Measurements of West Nile Virus in Wastewater Solids
WWSCAN Factsheets and Guidance Documents can be found here.
Wastewater Category Logic
As a reminder, WWSCAN categorizes sites based on Levels + Trends + Frequency of detection. We adopted this methodology back in 2023 as a way to clearly communicate what wastewater is telling us about community disease circulation. The wastewater categorization helps us quickly understand if the recent measurements for a infectious disease fall into a low, medium, or high category and are determined based on a combination of the following variables:
- Trends tell us how concentrations are changing (↑, ↓, -) over the last 21 days
- Levels tell us whether concentrations are relatively lower or higher (based on national tertiles over last 365 days)
- Frequency of detection indicates how often a pathogen is detected in wastewater (important for pathogens that are not commonly detected)
There are 3 Archetypes for WWSCAN Pathogens:
- Pathogens that are commonly detected (regardless of season - includes SARS-CoV-2, Norovirus)
- Seasonal pathogens (rarely detected outside of season/outbreaks - includes Influenza A + B, RSV, HMPV, EV-D68)
- Pathogens that are not commonly detected (rare regardless of season - includes C. auris, HepA, H5 influenza marker, Measles, Mpox clade Ib, Mpox clade II, West Nile Virus)
*Categories are not currently being calculated for H1/H3 Influenza markers, Parvovirus, Rotavirus or Adenovirus Group F.
To learn more about WWSCAN's categorization logic, please see: https://data.wastewaterscan.org/about/#8
COVID-19
SARS-CoV-2 concentration in wastewater and sequencing for variants
SARS-CoV-2 N gene RNA was detected in ~94% of samples in February 2026. The median SARS-CoV-2 concentration across all sites in February 2026 was 31,869 copies/gram. The median SARS-CoV-2 concentration in February 2026 was up ~11% compared to January 2026. While SARS-CoV-2 concentrations are very low in the West, we are continuing to see elevated concentrations across the Midwest and Northeast regions.
Below is the population weighted average SARS-CoV-2 N gene concentrations (normalized by PMMoV) is shown in black from all participating WWSCAN sites across the country over the last 365 days.
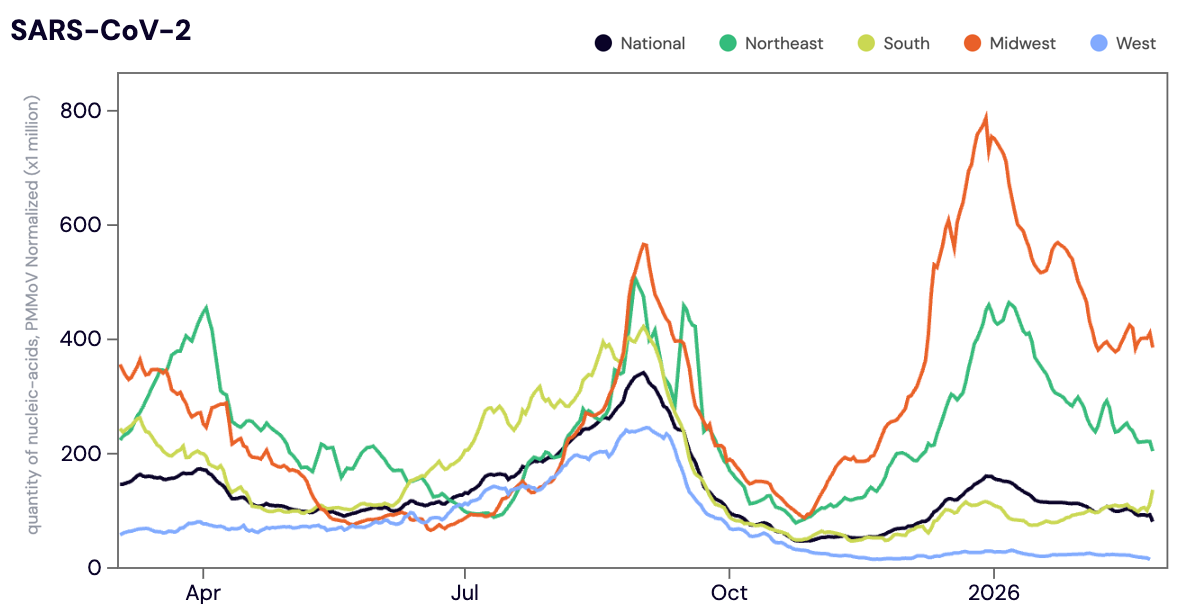
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, SARS-CoV-2 is in the 'HIGH' category with aggregated measurements showing high concentrations and a downward trend over the last 3 weeks.
Below are the current SARS-CoV-2 wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level).
- 56 are in the LOW Wastewater Category
- 29 are in the MEDIUM Wastewater Category
- 54 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
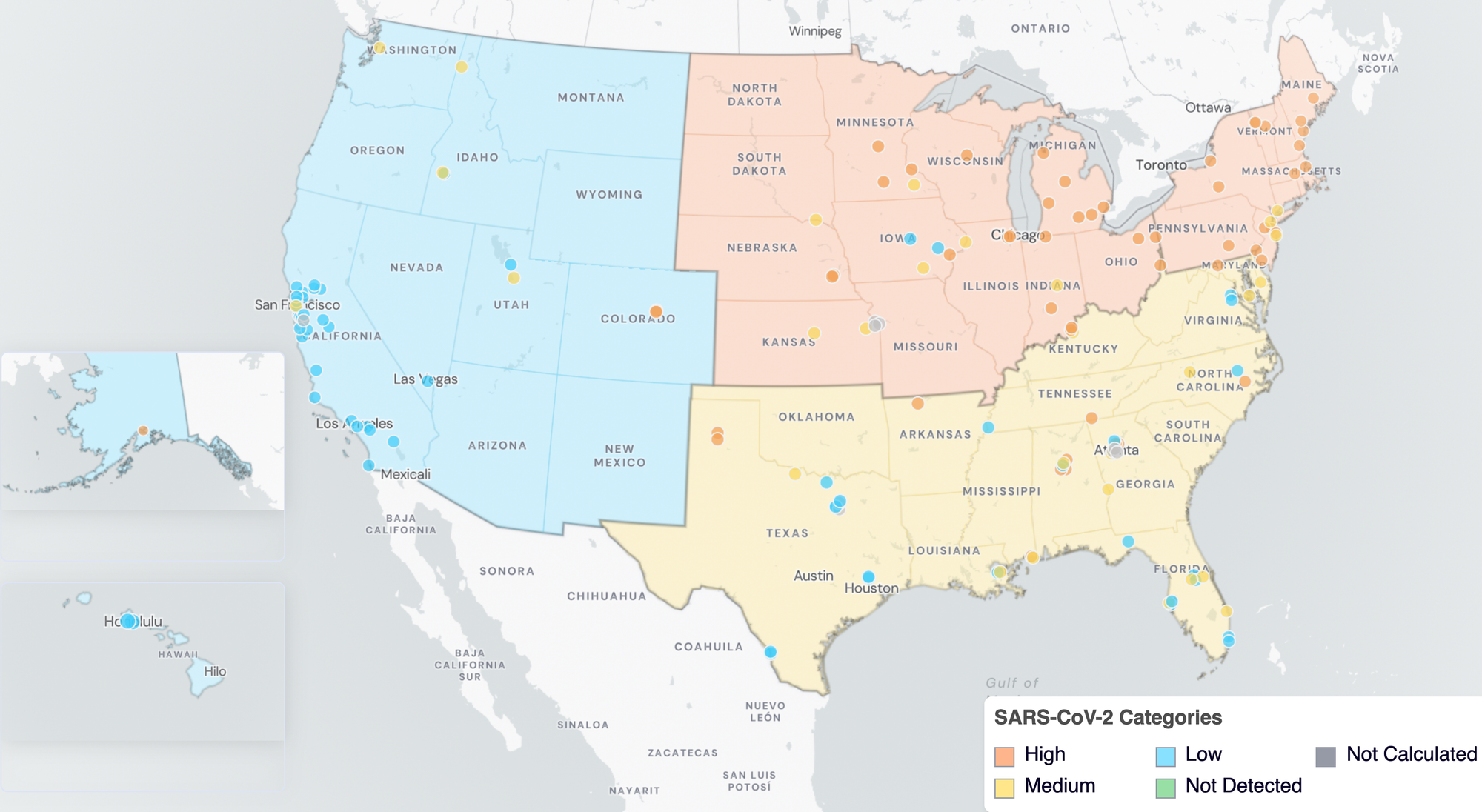
SARS-CoV-2 Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
Sequencing of SARS-CoV-2 from wastewater is available for a select number of individual sites. The sequencing protocol is available on protocol.io. The plots below show the relative proportions of different variants inferred from sequencing the entire genome of SARS-CoV-2. Results are based on sequencing of 2 samples per week, combined to provide a weekly value.
Sequencing data is also available in aggregate across all sites in WWSCAN with sequencing data. That plot is shown below and also suggests that XFG (shown in orange) is the most abundant variant across all sites at 71% followed by NB.1.8.1 (shown in mauve) at 6.8%, LF.7 (shown in yellow) at 4.9% and BA.3.2 (shown in blue) at 4.1%.
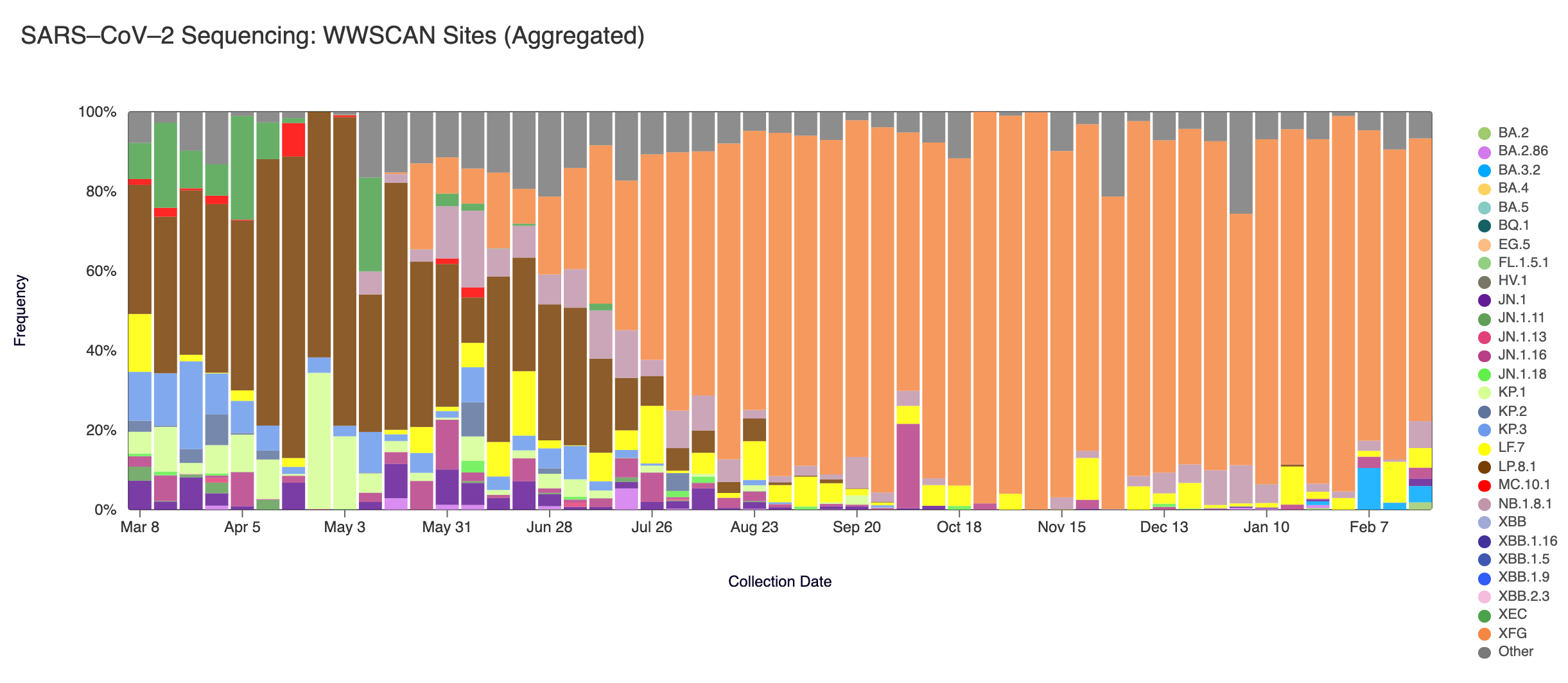
SARS-CoV-2 sequencing data from the last 12 months. This plot includes data through the week of February 21, 2026.
Other Respiratory Pathogens
Influenza A & B, Respiratory Syncytial Virus, Human metapneumovirus, EV-D68 & Parvovirus
Influenza A (IAV) RNA was detected in 95% of samples in February 2026. The median IAV RNA concentration across all sites in February 2026 was 22,571 copies/gram. The median IAV concentration in February 2026 was down ~36% compared to January 2026. While IAV concentrations are trending down, all regions are still in seasonal onset and majority of sites are still seeing elevated levels of IAV.
Below is the population weighted average IAV concentrations (normalized by PMMoV) is shown in black from all participating WWSCAN sites across the country over the last 365 days.
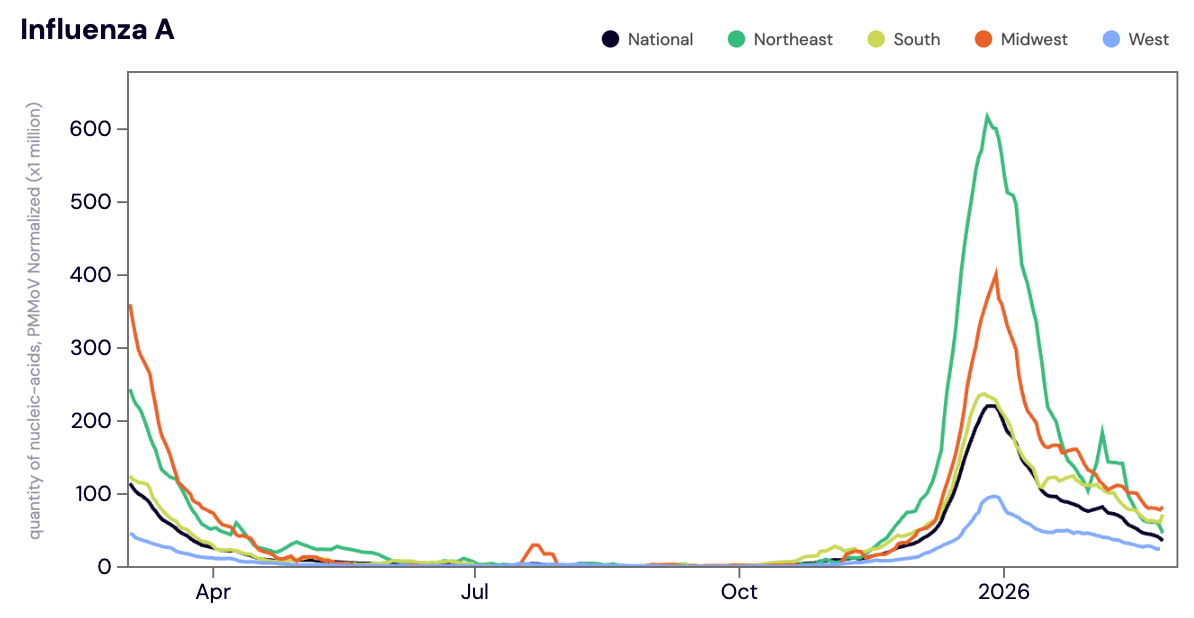
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, Influenza A is in the 'HIGH' Wastewater Category. We are seeing high concentrations and a downward trend for IAV in wastewater at the national level over the last 3 weeks.
Below are the current Influenza A wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level when in seasonal onset).
- 7 are in the LOW Wastewater Category
- 27 are in the MEDIUM Wastewater Category
- 105 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
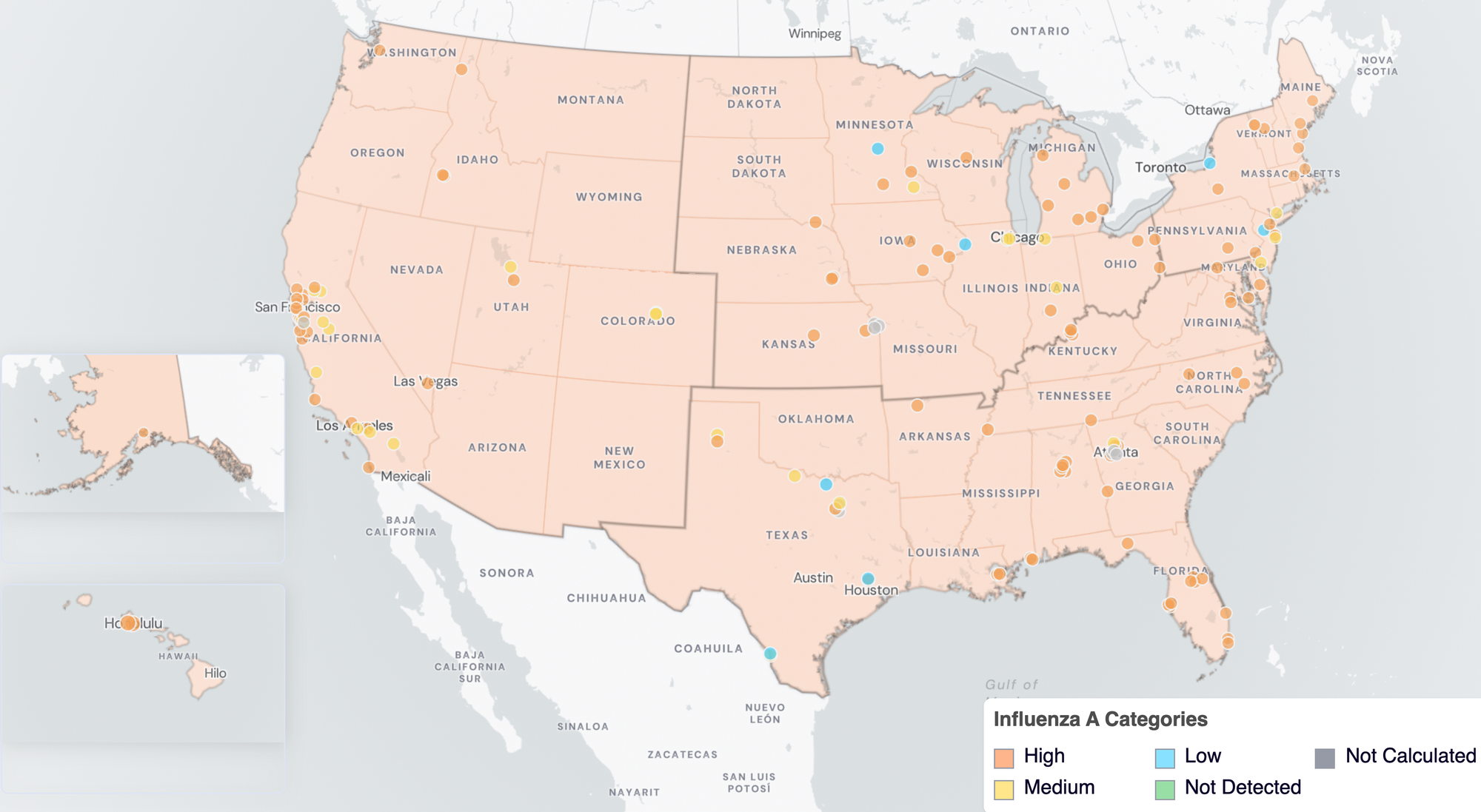
Influenza A Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
H1 Marker in Influenza A (IAV) The H1 marker in Influenza A was detected in ~31% of samples in February 2026. The H1 median concentration across all sites in February 2026 was below the limit of detection. You can find a heat map of positive H1 detections here. This flu season were are seeing very low detections of the H1 influenza marker compared to the H3 influenza marker.
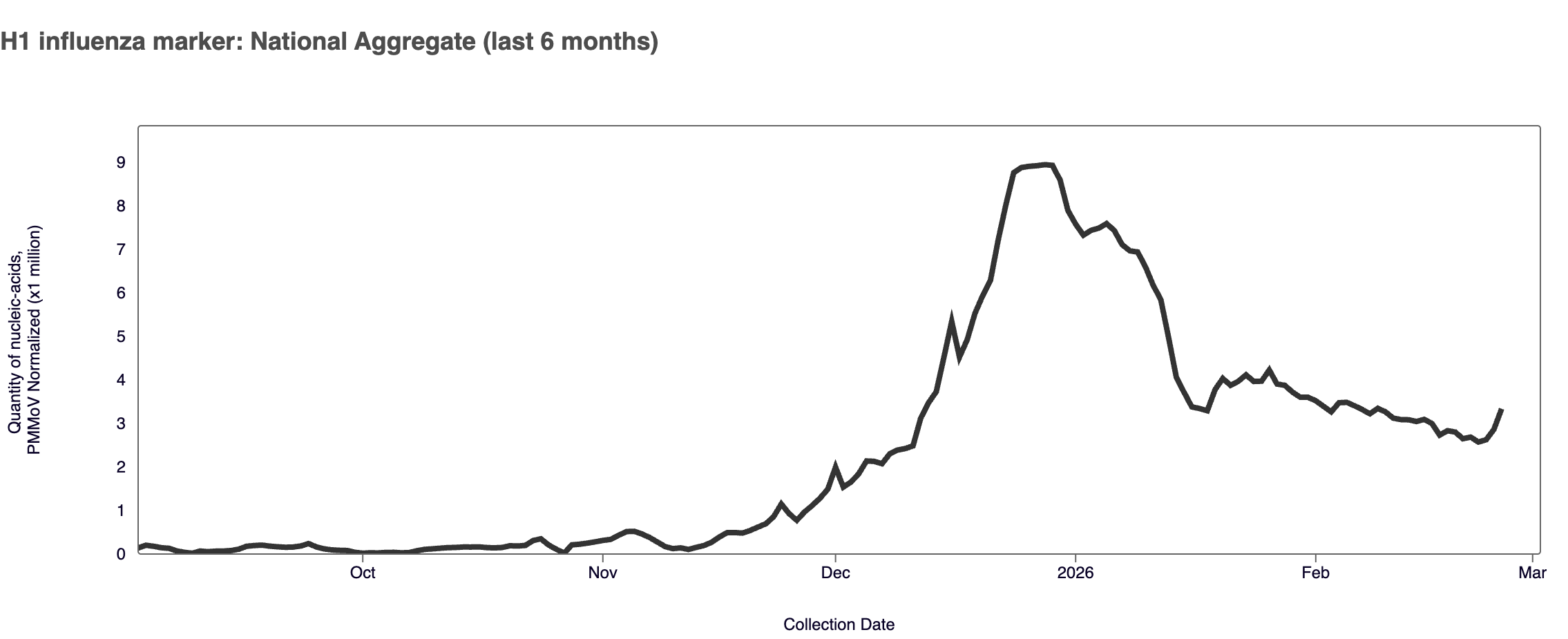
Plot shows last 6 months of data for the WWSCAN national aggregate of the H1 marker. Note the y-axis scale shows normalized wastewater concentrations for the H1 influenza marker.
H3 Marker in Influenza A (IAV) In February 2026, the H3 marker in influenza A was detected in ~87% of samples. The H3 marker median concentration across all sites in February 2026 was 12,515 copies per gram. The median H3 marker concentration in February 2026 was down ~42% compared to January 2026. At many plants, H3 marker concentrations are increasingly driving the higher influenza A concentrations. A detection of H3 could include subclade K, a highly-mutated H3N2 variant currently spreading worldwide. You can find a heat map of positive H3 detections here.
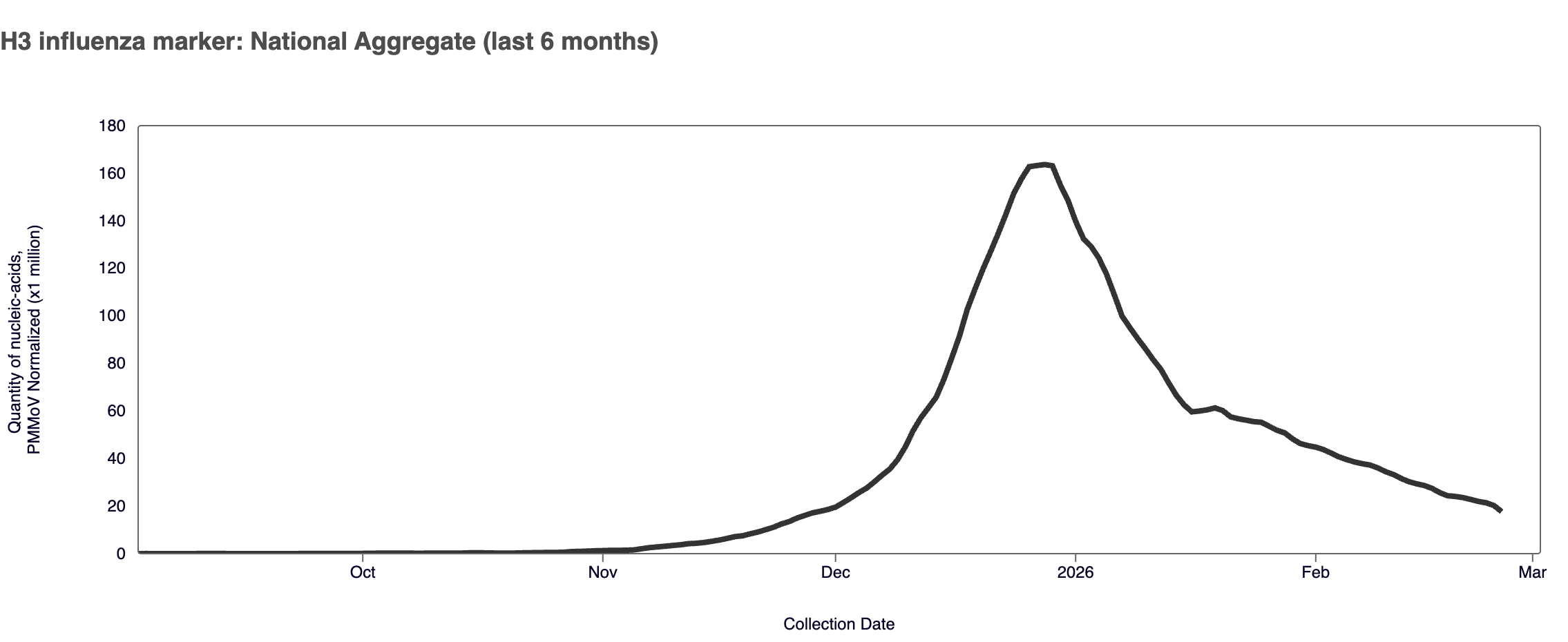
Plot shows last 6 months of data for the WWSCAN national aggregate. Note the y-axis scale shows normalized wastewater concentrations for the H3 influenza marker.
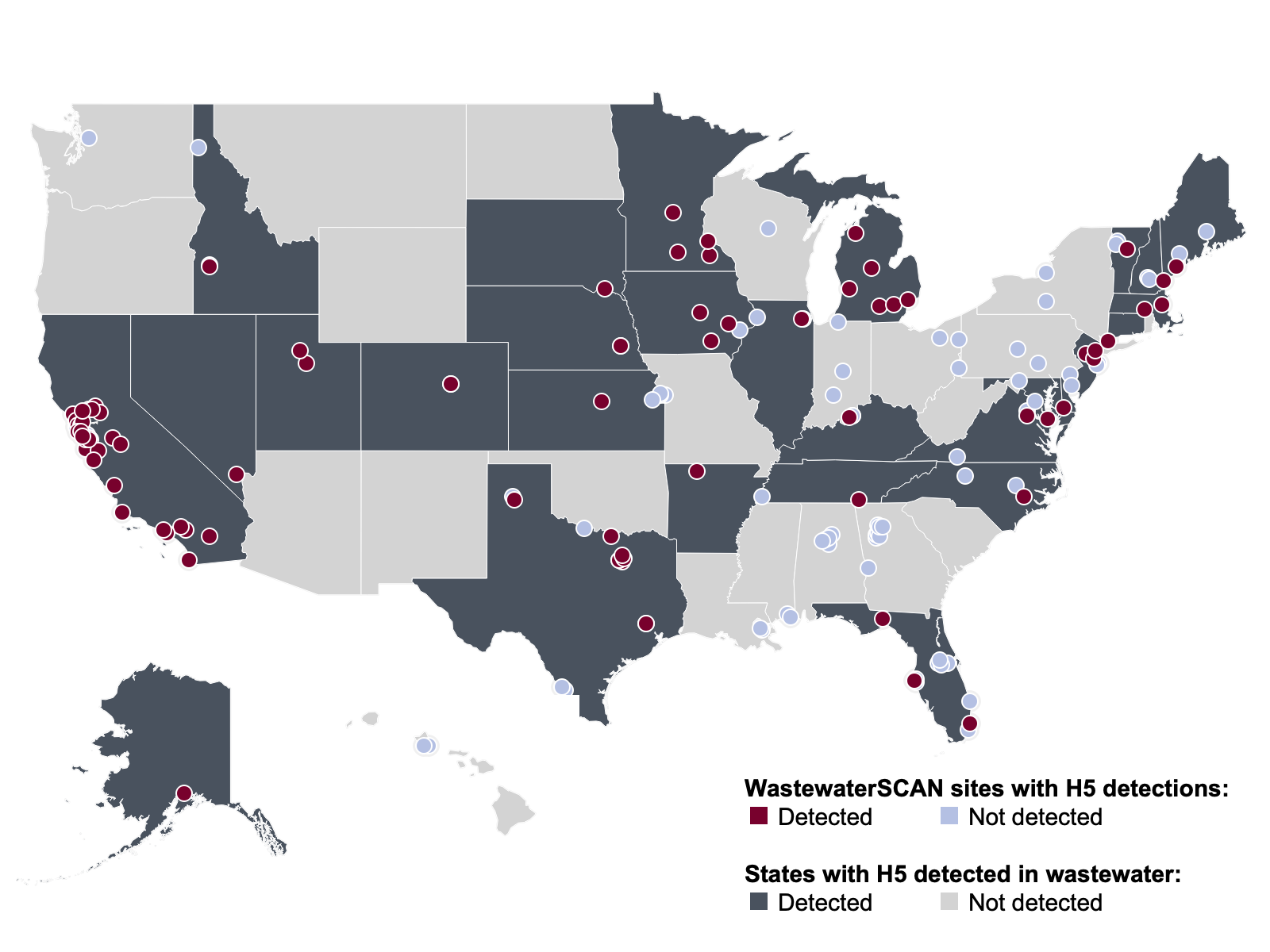
H5 Marker in Influenza A (IAV) As of February 27, 2026, WWSCAN sites across 28 states (AK, AR, CA, CO, CT, DE, FL, IA, ID, IL, KS, KY, MA, ME, MD, MI, MN, NC, NE, NH, NJ, NV, SD, TN, TX, UT, VA and VT) have seen positive detections for the H5 marker in influenza A. Since prospective monitoring began, 1,061/42,322 samples have tested positive (~2.5% detection rate). In February 2026, there were 8 positive H5 marker detection (1 detection in IN, and 7 detections in NJ). You can find a heat map of positive detections here.
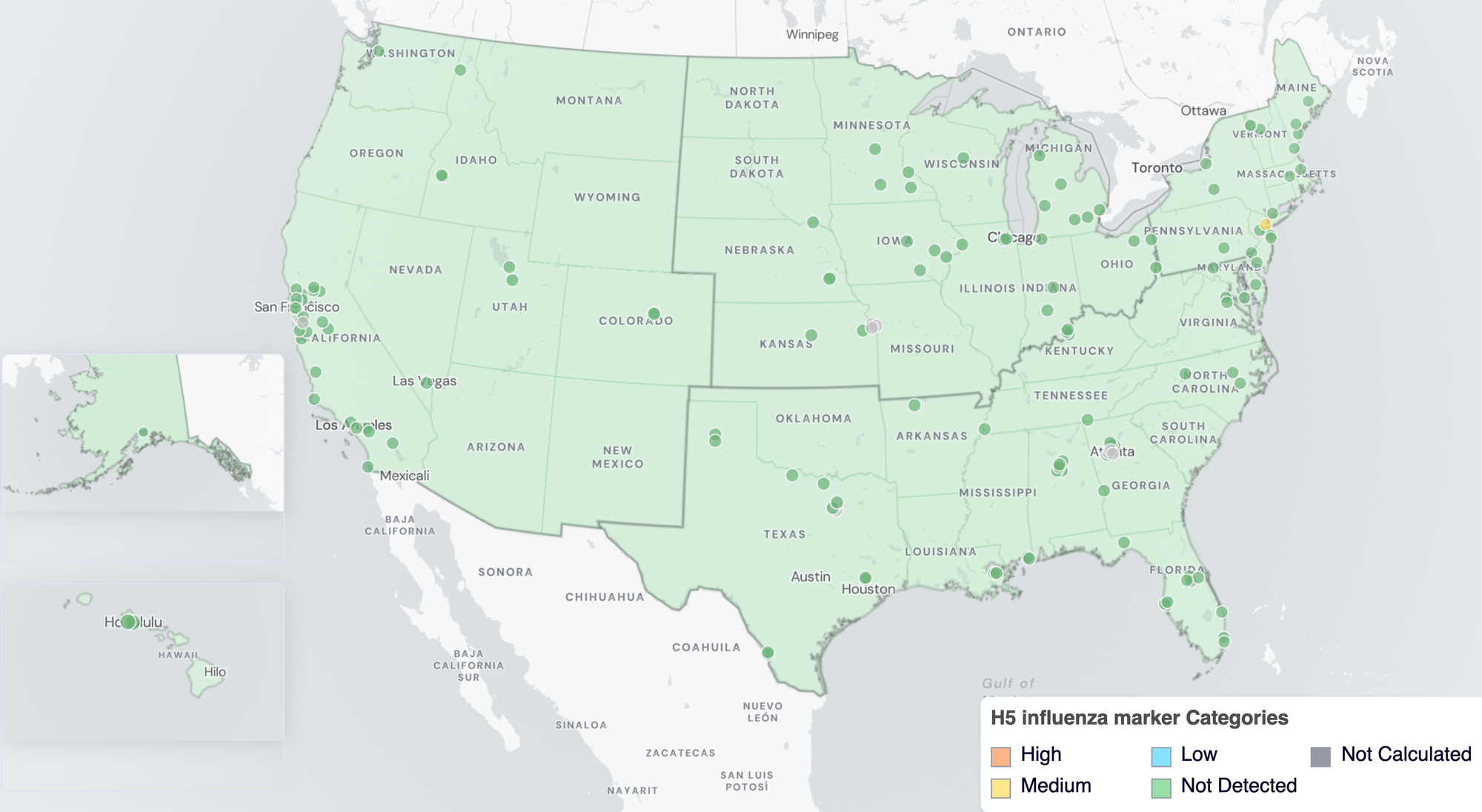
In January 2026 WWSCAN started calculating a wastewater category for the H5 influenza marker. The H5 marker is now assigned to the 'Pathogens that are not commonly detected ' category logic. Nationally, the H5 influenza marker is in the 'NOT-DETECTED' category due to no recent detections in the last 10 days.
Below are the H5 influenza marker wastewater categories for all WWSCAN sites (recall the category is determined by the frequency of detection).
- 138 are in the NOT-DETECTED Wastewater Category
- 0 are in the LOW Wastewater Category
- 1 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
H5 influenza marker Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
Influenza B (IBV) RNA was detected in ~74% of samples in February 2026. The median IBV RNA concentration across all sites in February 2026 was 8,021 copies/gram. For reference the median IBV concentration in January 2026 was below the limit of detection. Concentrations of IBV have been elevated in sites across the Midwest since early December 2025 and we've seen IBV concentrations steadily increasing across sites in the Northeast and South since late January 2026. As a reminder, during last years respiratory virus season (2024-2025) the national WWSCAN network did not hit seasonal onset for IBV until early March 2025.
Below is the population-weighted average IBV RNA concentrations (normalized by PMMoV) is shown in black from all participating WWSCAN sites across the country over the last 365 days.
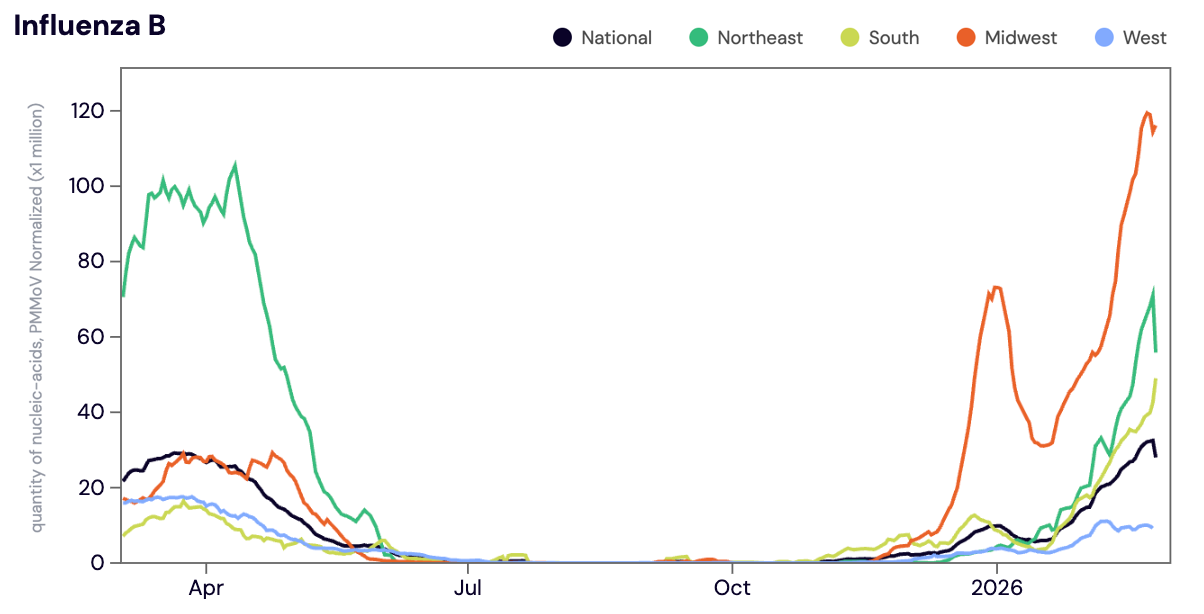
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, Influenza B is in the 'HIGH' Wastewater Category due to >50% of sites being in onset for IBV in wastewater. We are seeing high concentrations and a upward trend for IBV in wastewater at the national level over the last 3 weeks.
Below are the current Influenza B wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level when in seasonal onset).
- 58 are in the LOW Wastewater Category
- 0 are in the MEDIUM Wastewater Category
- 81 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
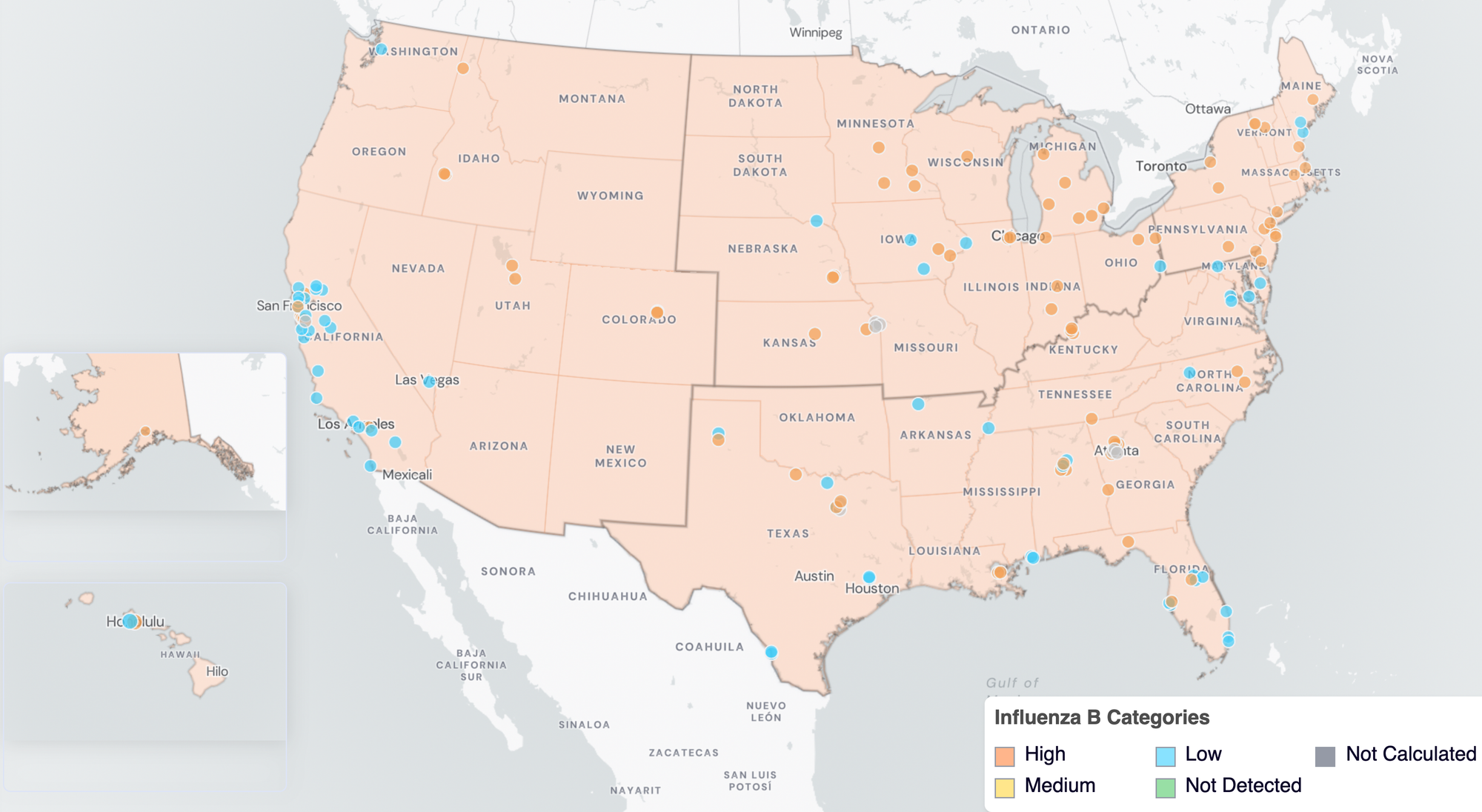
IBV Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
Respiratory syncytial virus (RSV) RNA was detected in ~92% of samples in February 2026. The RSV RNA median concentration across all sites in February 2026 was 20,409 copies per gram. The median RSV concentration in February 2026 was up ~29% compared to January 2026. As a reminder, during last years respiratory virus season the WWSCAN network hit seasonal onset for RSV in early December 2024 and peaked in late January/early February 2025. We've seen a slower start to the RSV wastewater wave this year (2025-2026) compared to last year (2024-2025).
Below the population-weighted average RSV RNA concentrations (normalized by PMMoV) is shown in black from all participating WWSCAN sites across the country over the last 365 days.
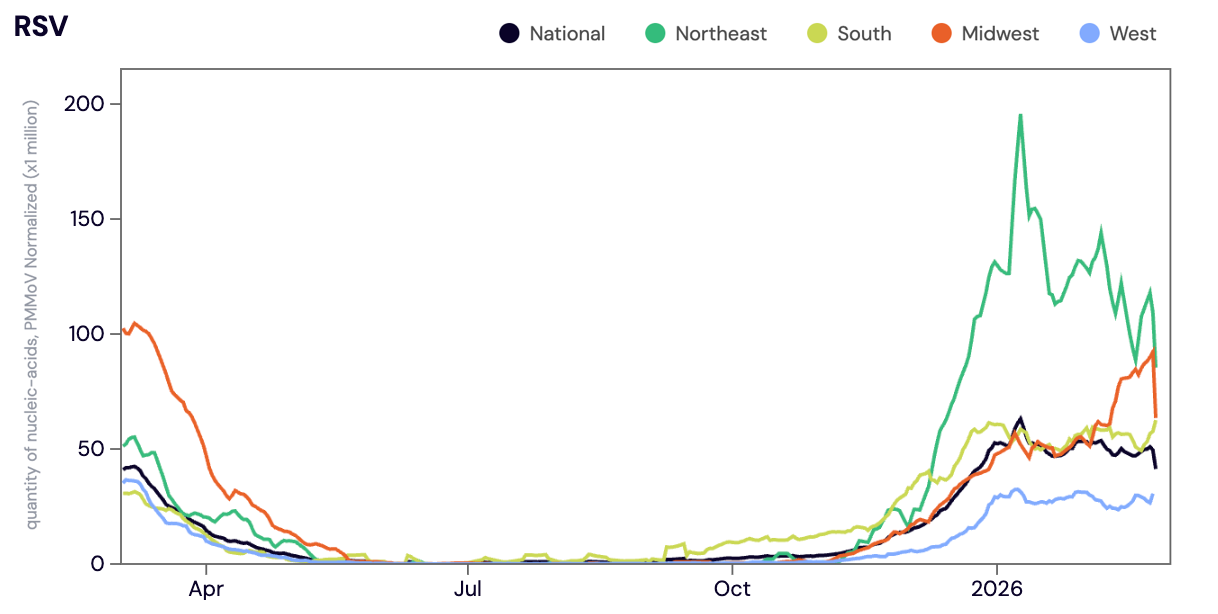
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, RSV is in the 'HIGH' Wastewater Category with >50% of sites in seasonal onset for RSV in wastewater. We are seeing high concentrations and a upward trend for RSV in wastewater at the national level.
Below are the current RSV wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level when in seasonal onset).
- 14 are in the LOW Wastewater Category
- 5 are in the MEDIUM Wastewater Category
- 120 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
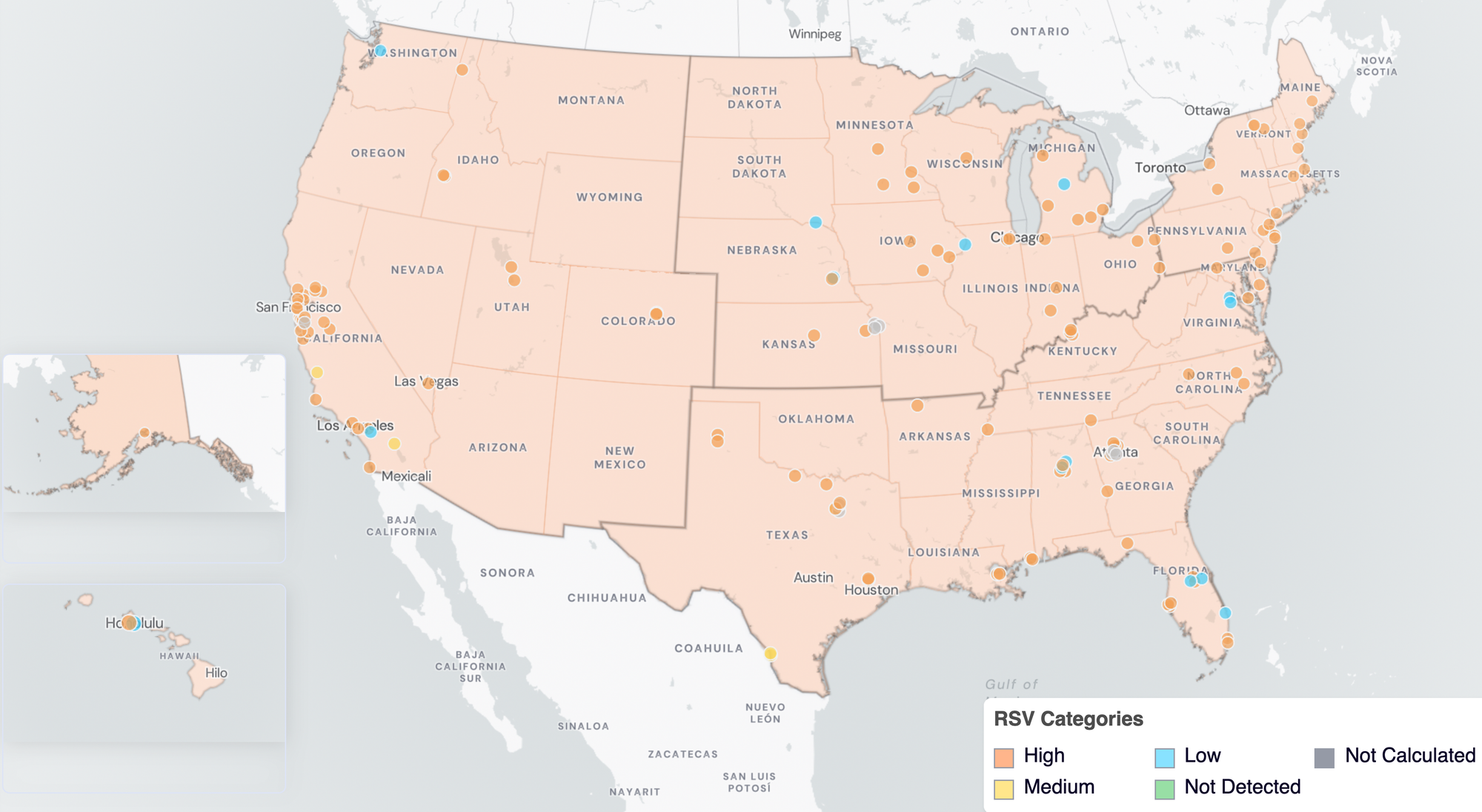
RSV Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
HMPV (human metapneumovirus) RNA was detected in ~71% of samples in February 2026. The median HMPV RNA concentrations across all sites in February 2026 was 8,630 copies per gram. The median HMPV concentration in February 2026 was up ~70% compared to January 2026. HMPV concentrations have been steadily rising across all regions since late December 2025.
Below is the population weighted average HMPV RNA concentrations (normalized by PMMoV) shown in black from all participating WWSCAN sites across the country over the last 365 days.
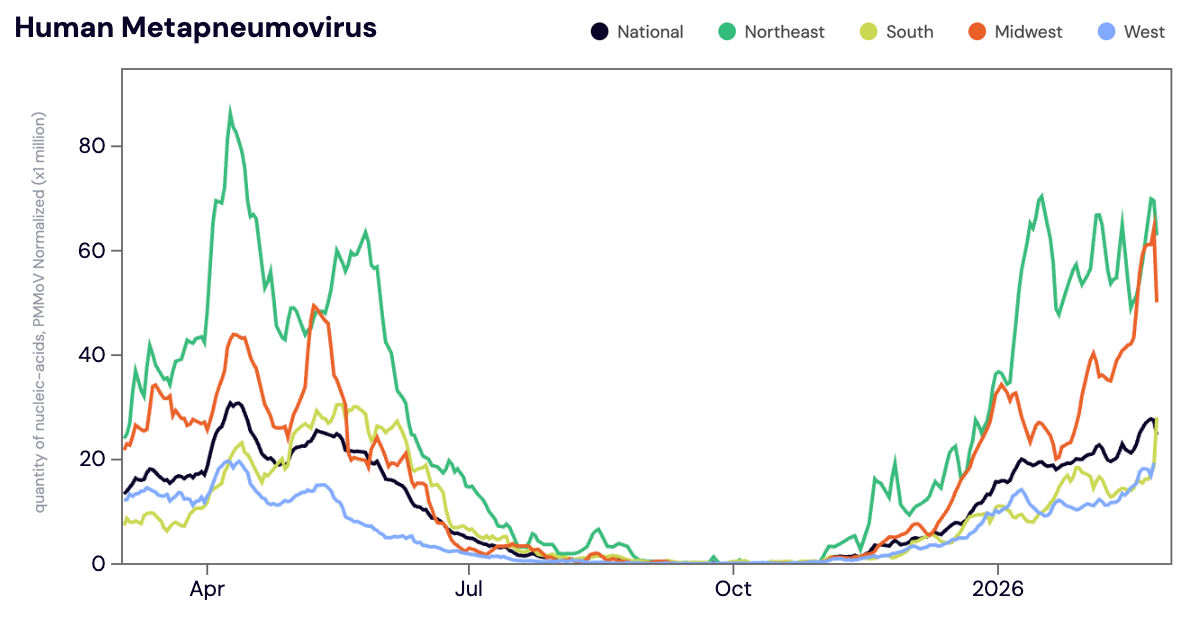
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, HMPV is in the 'HIGH' wastewater category as >50% of sites in seasonal onset for HMPV in wastewater. We are seeing high concentrations and a upward trend for HMPV in wastewater at the national level.
Below are the current HMPV wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level).
- 53 are in the LOW Wastewater Category
- 8 are in the MEDIUM Wastewater Category
- 78 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
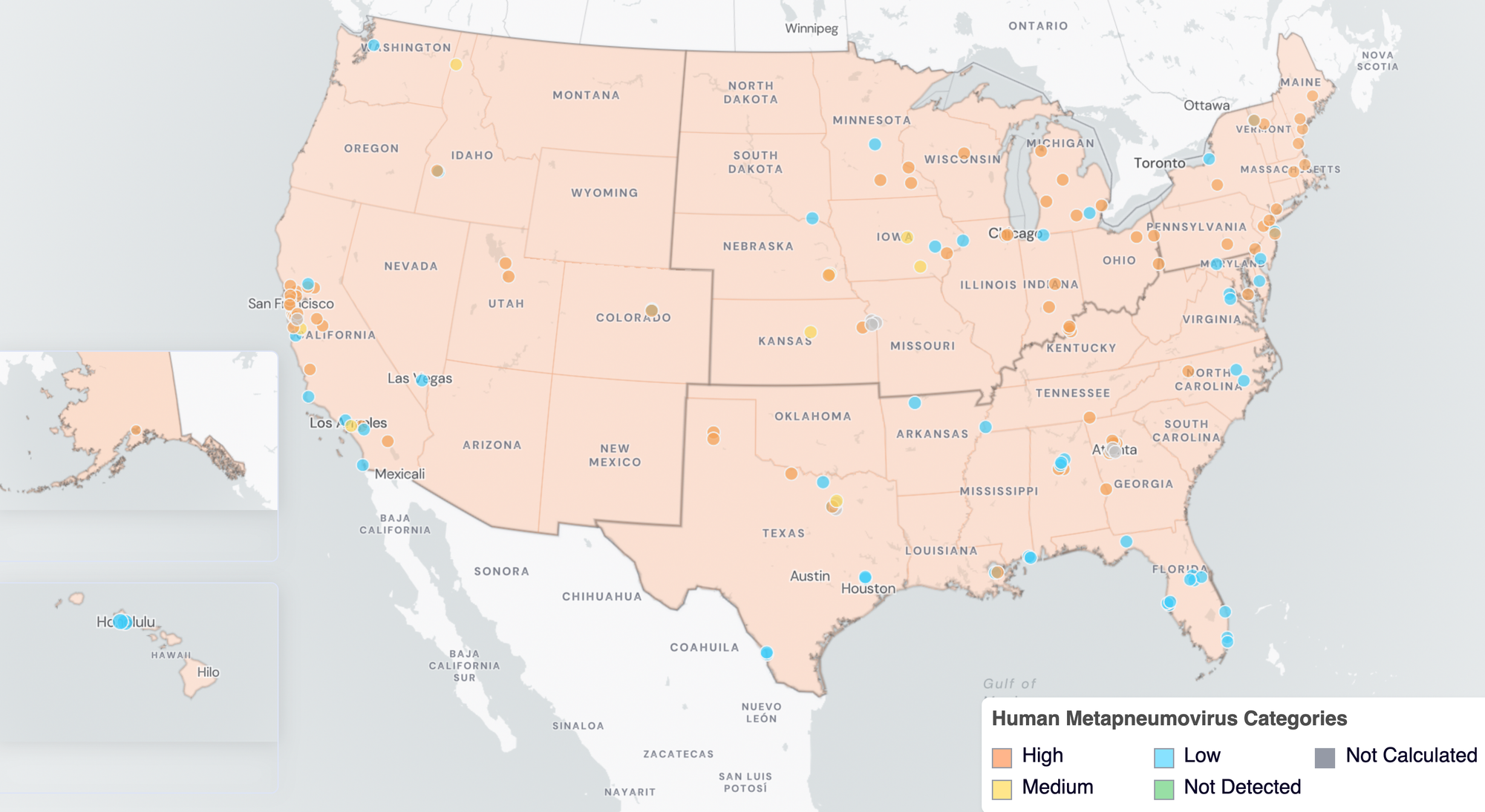
HMPV Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
Enterovirus D68 (EV-D68) RNA was detected in ~9% of samples collected in February 2026 (with most positive detections in California). The median concentration across all sites in February 2026 was below the limit of detection.
Below is the population-weighted average EV-D68 RNA (normalized by PMMoV) shown in black from all participating WWSCAN sites across the country over the last 24 months. In this plot you can see elevated EV-D68 concentrations across all regions during the 2024-2025 season. EV-D68 concentrations in the Midwest (shown in red) rose again in mid August 2025 before coming down in November 2025, and concentrations in the West (shown in blue) rose in late September 2025 and have come down since the beginning of January 2026.
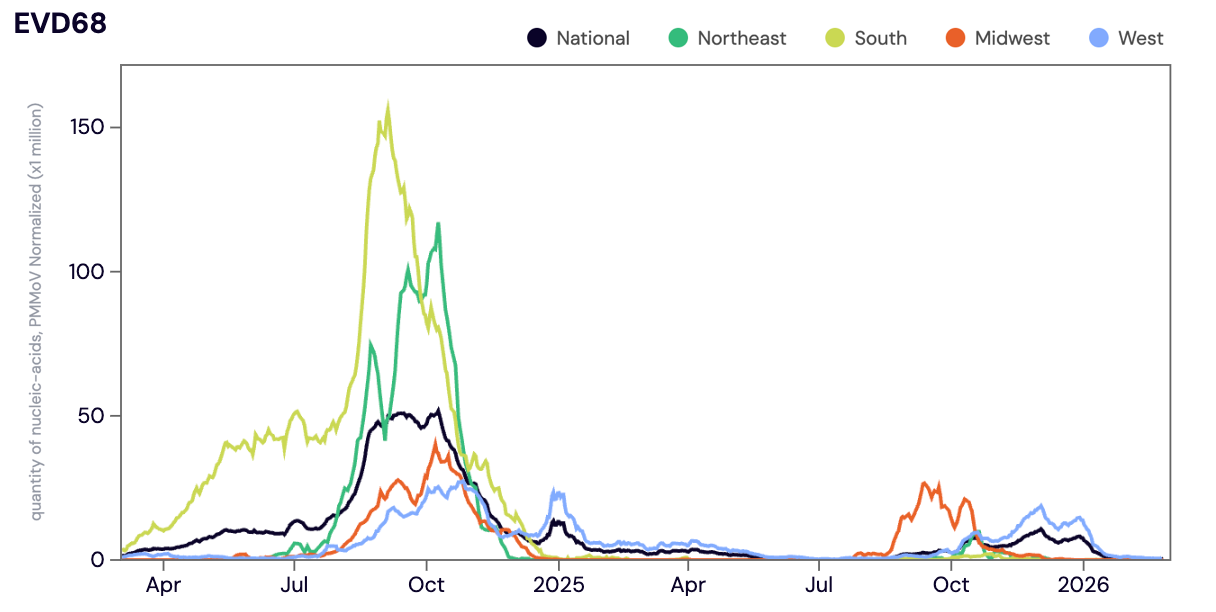
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, EV-D68 is in the 'LOW' Wastewater Category with >50% of sites in offset of EV-D68 in wastewater.
Below are the current EV-D68 wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level).
- 139 are in the LOW Wastewater Category
- 0 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
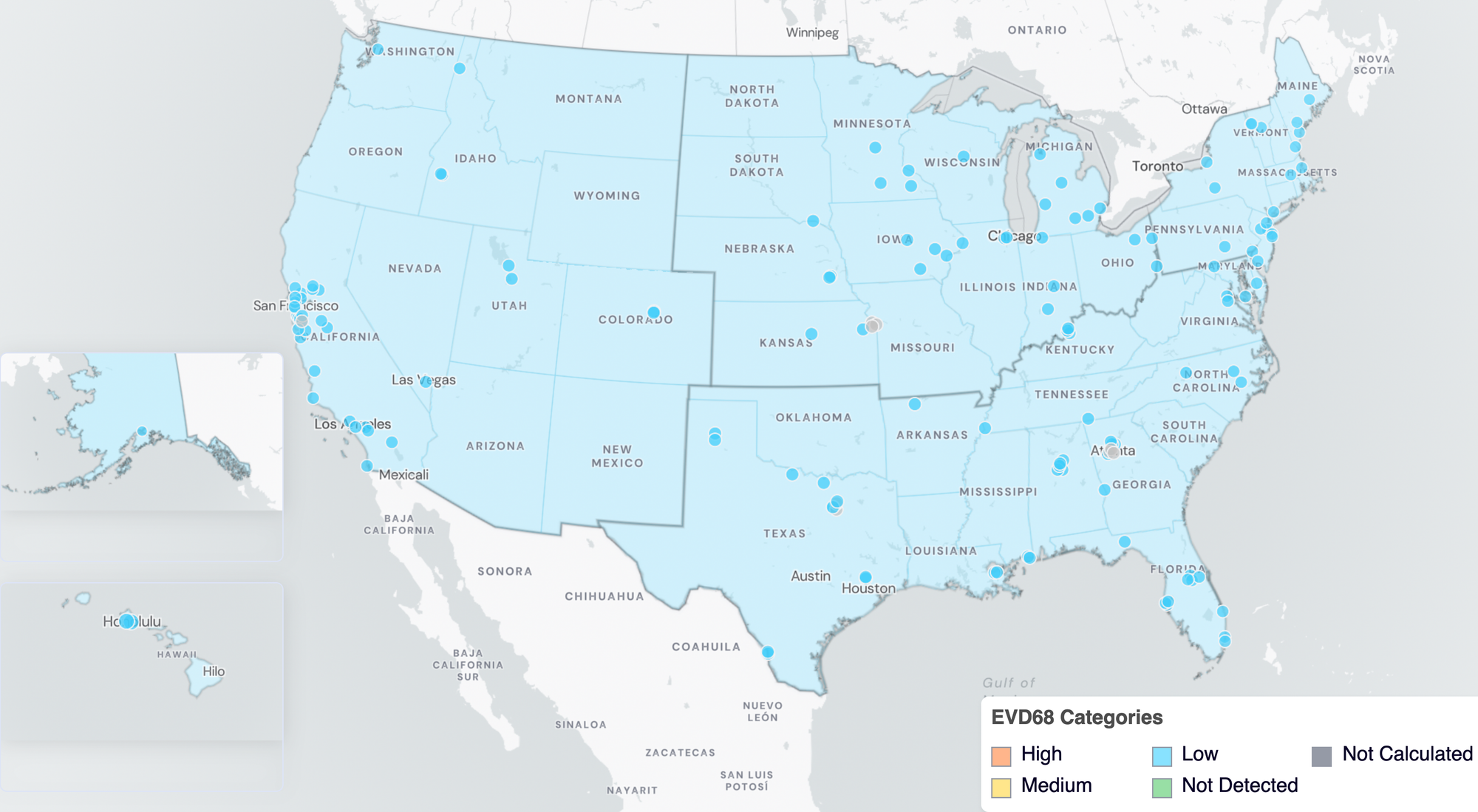
EV-D68 Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
Parvovirus (Parvo) DNA - In mid May 2025, WWSCAN began prospectively monitoring for Parvovirus. Parvovirus B19 DNA was detected in ~25% of samples collected in February 2026 across WWSCAN sites.
The chart below shows each WWSCAN plant in the Greater SF Bay Area as rows, and days as columns (labeled at the bottom axis). White indicates no samples, blue is a non-detect for Parvo DNA and red is a detection for Parvo DNA. Evident in the plot is sparse detections at some sites and frequent detections at other sites.
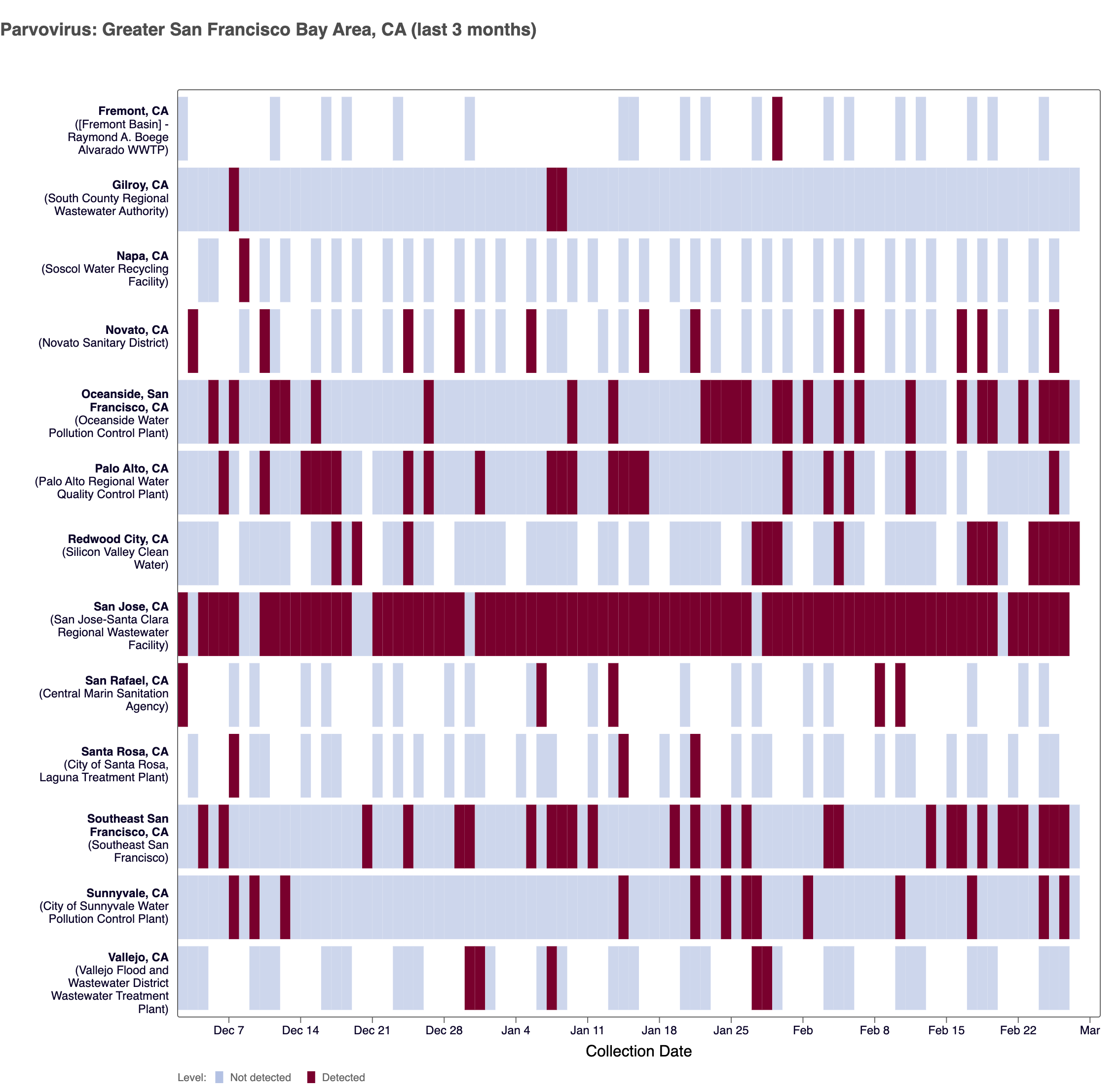
Heat map of positive Parvovirus detections (in red) across sites in the Greater SF Bay area participating in WWSCAN over the last 3 months. Blue indicates non-detect.
As a reminder, we are not currently calculating wastewater categories for Parvovirus. In order to calculate wastewater categories we need a longer time series of data to examine trends and levels.
We published a paper on 'Community Infections Linked with Parvovirus B19 Genomic DNA in Wastewater, Texas, USA, 2023–2024'. This paper covers retrospective analysis conducted at two WWTPs in Texas prior to WWSCAN rolling out the assay to all participating WWSCAN sites. You can find the paper linked here.
Gastrointestinal Pathogens
Norovirus GII, Human Adenovirus Group F, & Rotavirus
Norovirus GII (HuNoV GII) RNA was detected in 100% of samples analyzed in February 2026. The median Norovirus concentration in February 2026 was up ~44% compared to January 2025. Last year, we saw Norovirus concentrations start to increase in early October 2024 and peak early February 2025. While we have seen a slower start to the winter Norovirus wave this year (2025-2026), the pattern is similar to what we observed during the 2023-2024 winter Norovirus wave.
Below is the population weighted average HuNoV GII RNA concentrations (normalized by PMMoV) shown in black from all participating WWSCAN sites across the country over the last 24 months.
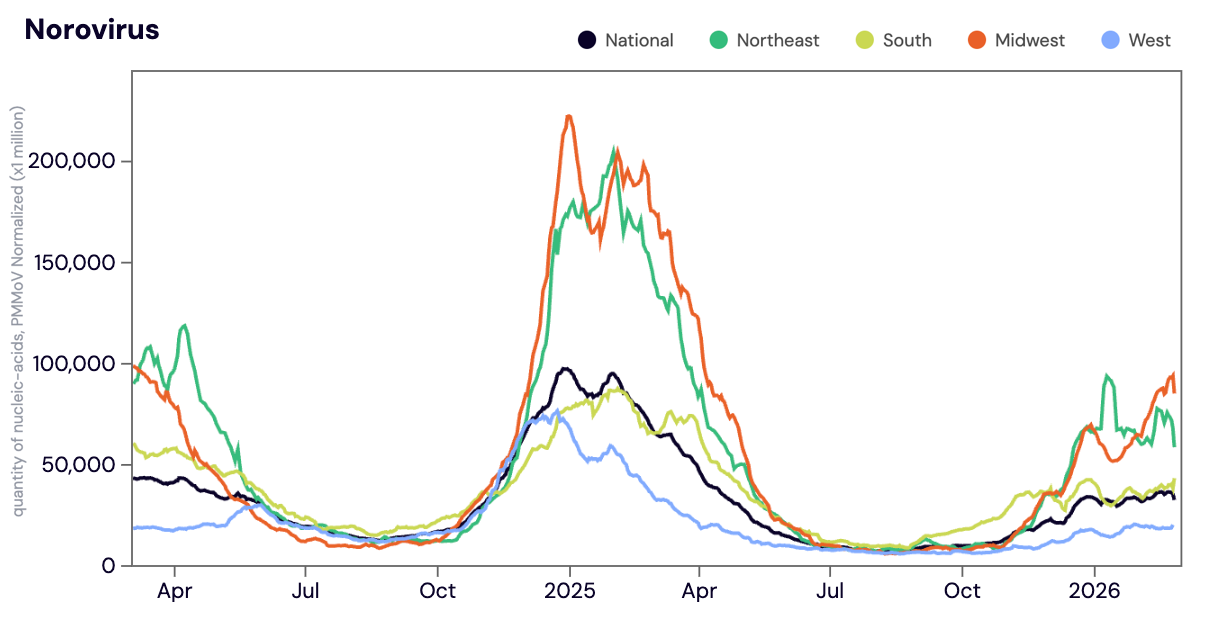
Midwest states: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast states: CT, MA, ME, NH, NJ, NY, PA, VT. Southern states: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western States: AK, CA, CO, HI, ID, NV, UT, WA.
Nationally, Norovirus is in the 'HIGH' Wastewater Category with aggregated measurements showing high concentrations and no trend in the last 21 days. The Midwest, Northeast and South are in the 'HIGH' wastewater category for Norovirus, while the West is in the 'MEDIUM' category.
Below are the current Norovirus wastewater categories for all WWSCAN sites (recall the category is determined by both the trend and the level).
- 14 are in the LOW Wastewater Category
- 31 are in the MEDIUM Wastewater Category
- 94 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
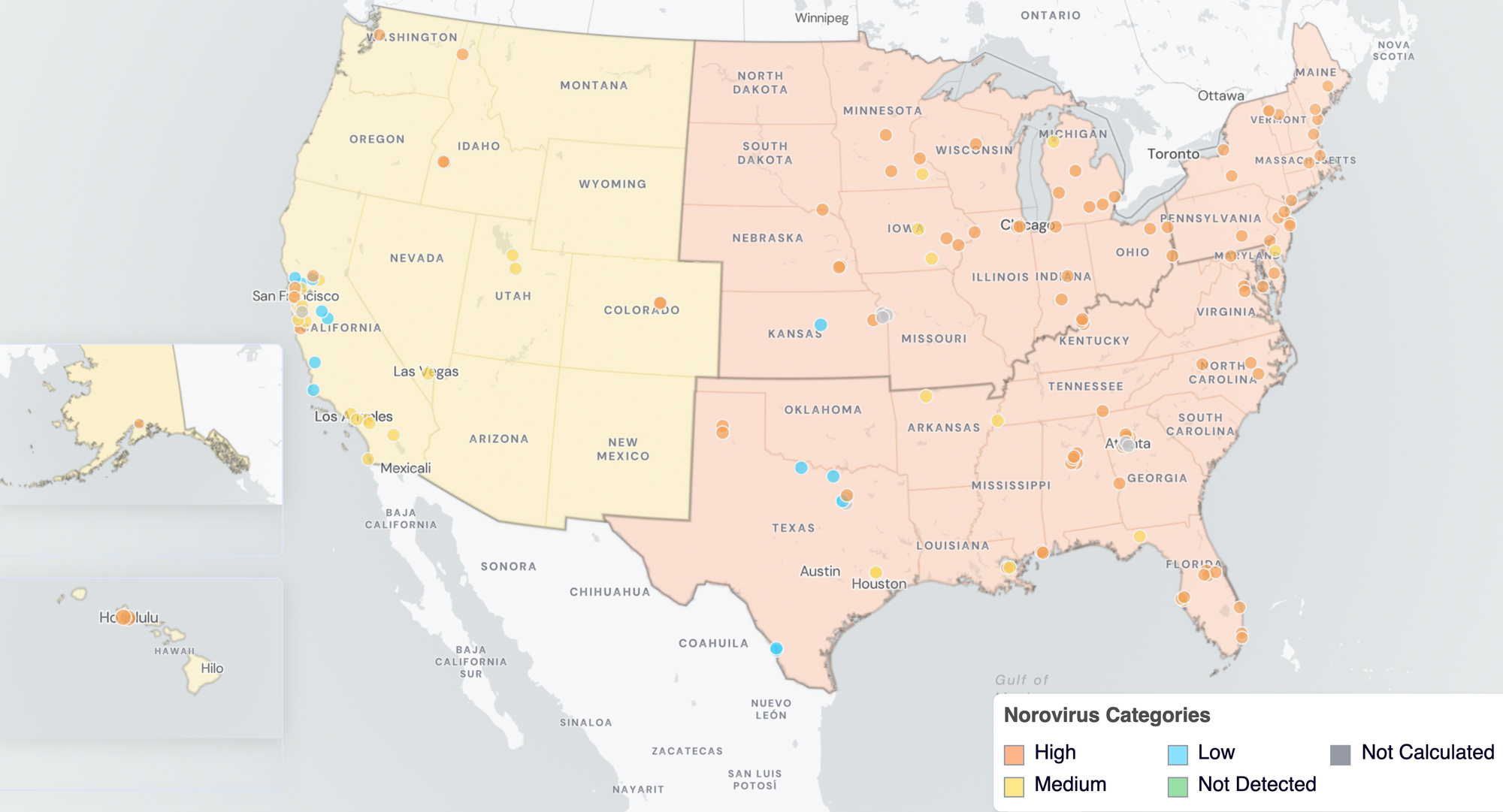
Norovirus Wastewater Categories. This map includes sample collection data through February 27, 2026. Each dot represents a single site and the four US Census regions are outlined in gray. Midwest Region includes: IA, IL, IN, KS, MI, MN, NE, OH, SD, WI. Northeast Region includes: CT, MA, ME, NH, NJ, NY, PA, VT. Southern Region includes: AL, AR, DE, FL, GA, KY, LA, MD, MS, NC, TN, TX, VA, and WV. Western Region includes: AK, CA, CO, HI, ID, NV, UT, WA.
Human Adenovirus Group F (HAdV-F) RNA - In mid May 2025, WWSCAN began prospectively monitoring for HAdV-F.
HAdV-F RNA was detected in 100% of samples from all WWSCAN sites in January 2026. The median concentrations across all sites in February 2026 was 4,431,971 copies/gram, down ~10% from January 2025.
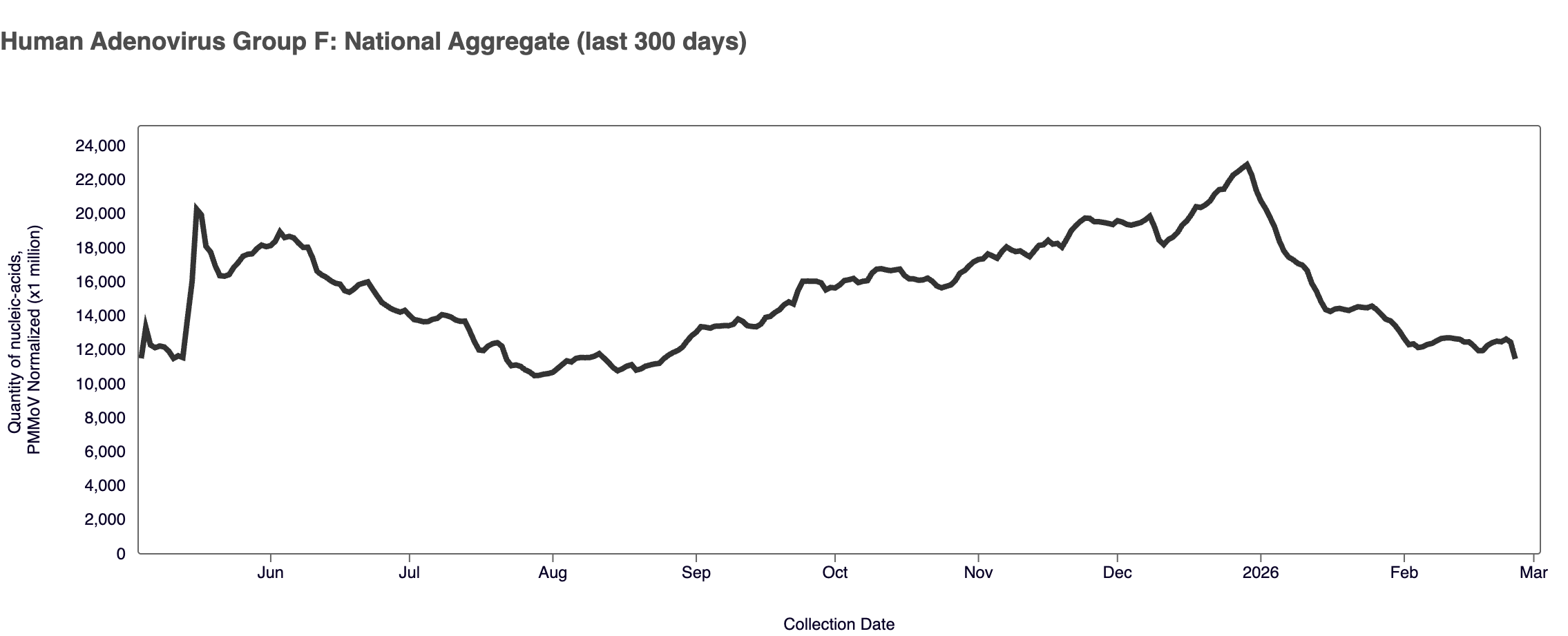
Plot shows ~ last 9 months of HAdV-F for the WWSCAN national aggregate since monitoring resumed. Note the y-axis scale shows normalized wastewater concentrations for HAdV-F.
As a reminder, we are not currently calculating wastewater categories for HAdV-F. In order to calculate wastewater categories we need a longer time series of data to examine trends and levels.
Rotavirus (Rota) RNA- In mid May 2025, we started measuring Rotavirus again. We previously paused measurements of Rota in December 2024 in order to being measuring Mpox clade Ib.
Rota RNA was detected in ~96% of samples from all WWSCAN sites in February 2026. The median concentrations across all sites in February 2026 was 103,779 copies/gram, up ~42% from January 2026.
Below is the is the population weighted average Rota RNA concentrations (normalized by PMMoV) shown in black from all participating WWSCAN sites across the country over the last 24 months. The dashed black lines indicate the period of time the Rota assay was paused (~6 months).
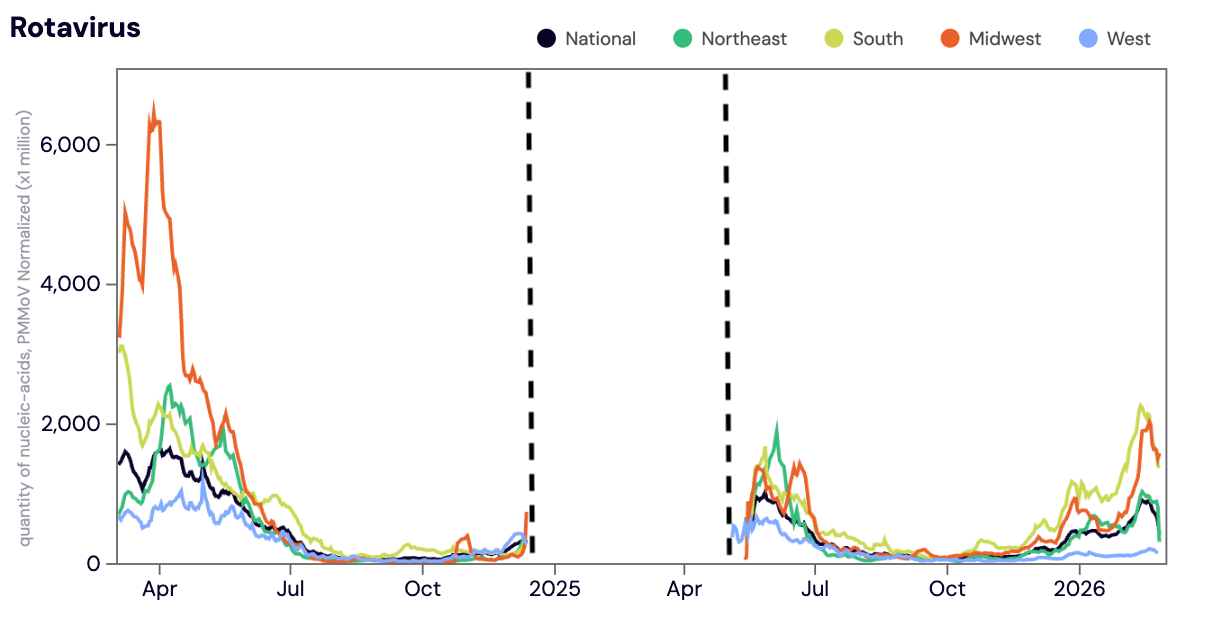
The dashed black lines indicate the period of time the Rota assay was paused (~6 months).
As a reminder, we are not currently calculating wastewater categories for Rota. In order to calculate wastewater categories we need a longer time series of data to examine trends and levels.
Other Pathogens of Concern
Candidozyma auris, Hepatitis A, Measles, Mpox clade Ib, Mpox clade II, and West Nile Virus
Candidozyma auris (C.auris) DNA - We detected C. auris DNA in ~3% of samples from all WWSCAN sites in February 2026. Nationally, C. auris is in the 'LOW' category due to limited detections.
The chart below shows plants in HHS Region 5 (IL, IN, MI, MN, OH, WI) as rows, and days as columns (labeled at the bottom axis). White indicates no samples, blue is non-detect for C. auris DNA and red is detect for C. auris DNA. Evident in the plot below is some sites have frequent detections while most have few to no detections of C. auris.
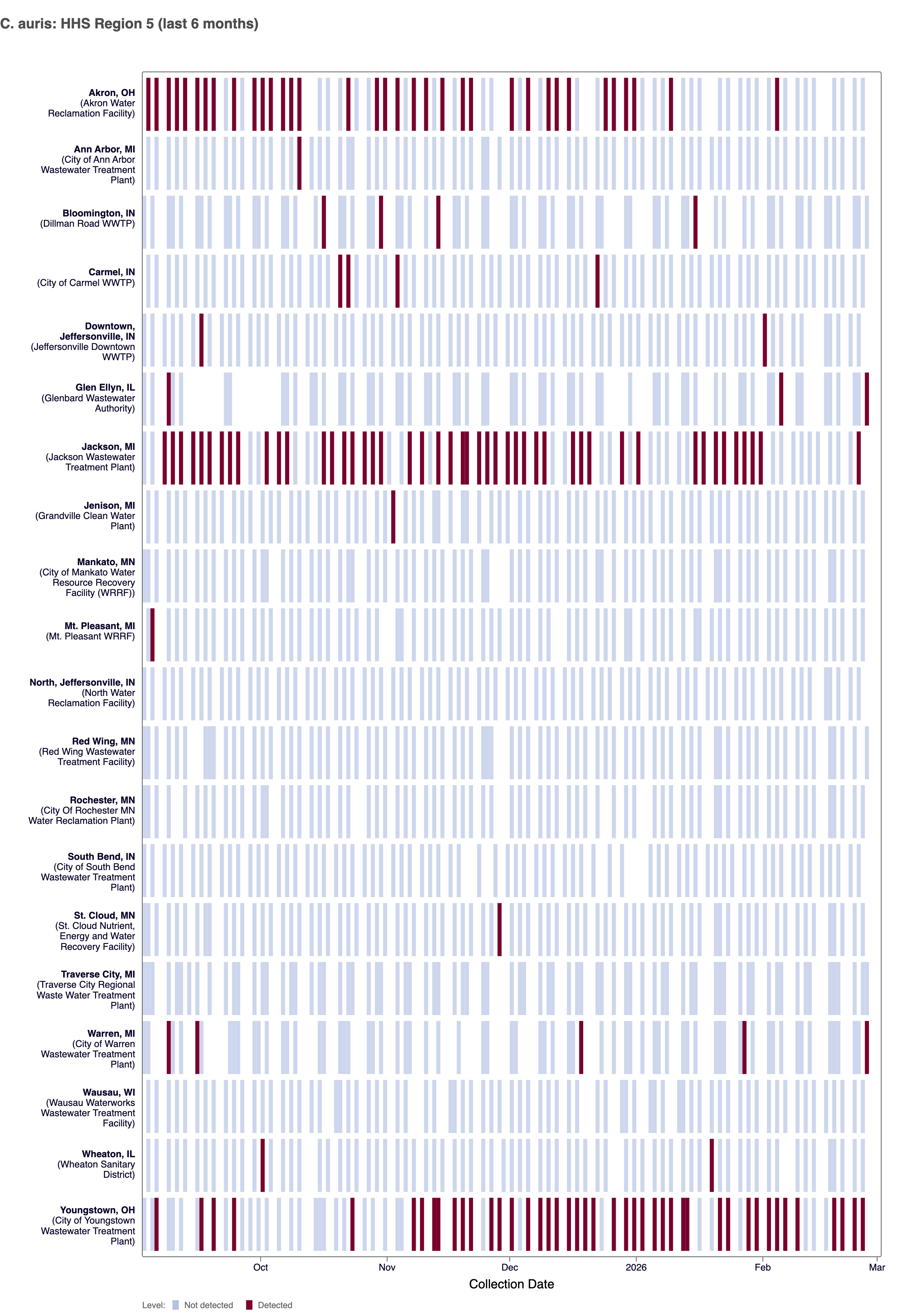
Heat map shows last 6 months of C. auris wastewater data from WWSCAN sites in HHS Region 5 (IL, IN, MI, MN, OH, WI). Blue indicates non-detect.
Below are the C. auris wastewater categories for all WWSCAN sites (recall the category is determined by the frequency of detection).
- 128 are in the NOT-DETECTED Wastewater Category
- 8 are in the LOW Wastewater Category
- 3 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
Hepatitis A (Hep A) RNA - We detected Hep A RNA in ~16% of samples from all WWSCAN sites in February 2026. Nationally, HepA is in 'LOW' category due to limited detections.
The chart below shows each WWSCAN plant in New Jersey as rows, and days as columns (labeled at the bottom axis). White indicates no samples, blue is a non-detect for Hep A RNA and red is a detection for Hep A RNA. Evident in the plot below is some sites have consistent detections while others have few to no detections of HepA.
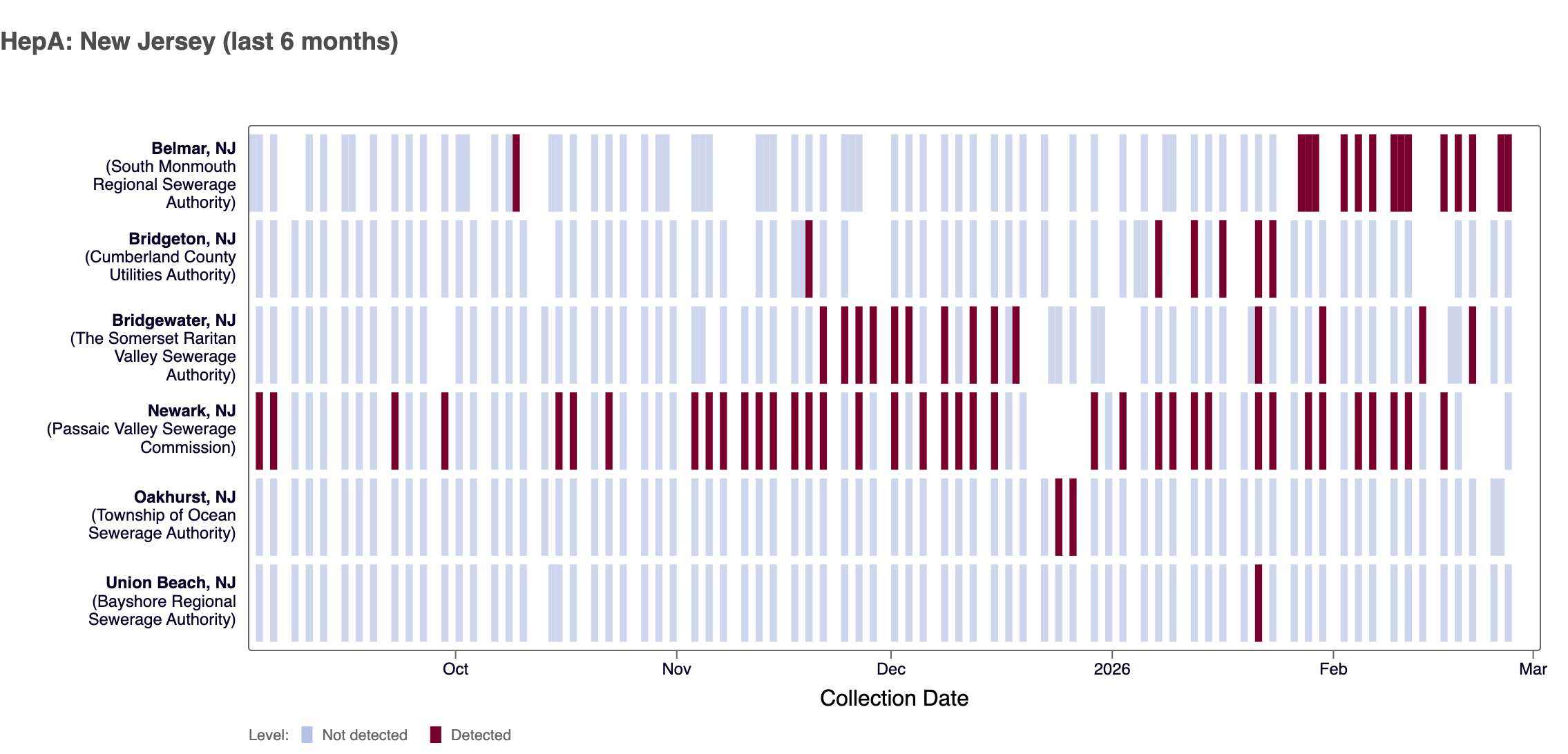
Heat map shows last 6 months of Hepatitis A wastewater data from WWSCAN sites in NJ. Blue indicates non-detect.
Below are the current HepA wastewater categories for all WWSCAN sites (the category is determined by the frequency of detection).
- 85 are in the NOT-DETECTED Wastewater Category
- 38 are in the LOW Wastewater Category
- 16 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
Measles (MeV) RNA - In mid-May 2025, WWSCAN began prospectively monitoring for MeV. A. Since prospective monitoring began, 121/18,538 samples have tested positive (~0.6% detection rate). In February 2026, there were 20 positive detections of MeV in wastewater samples (4 in CT, 2 in FL, 10 in UT, and 4 in VT). Nationally, MeV is in the 'LOW' category due to limited detections.
Since monitoring began, 37 sites across 20 states (AK, CA, CO, CT, FL, GA, IA, ID, IL, IN, KS, MD, MI, MN, NE, NV, NY, TX, UT and VT) have had at least 1 positive detection of MeV in wastewater samples collected. Below is a map of sites with MeV detections in wastewater since prospective monitoring began.
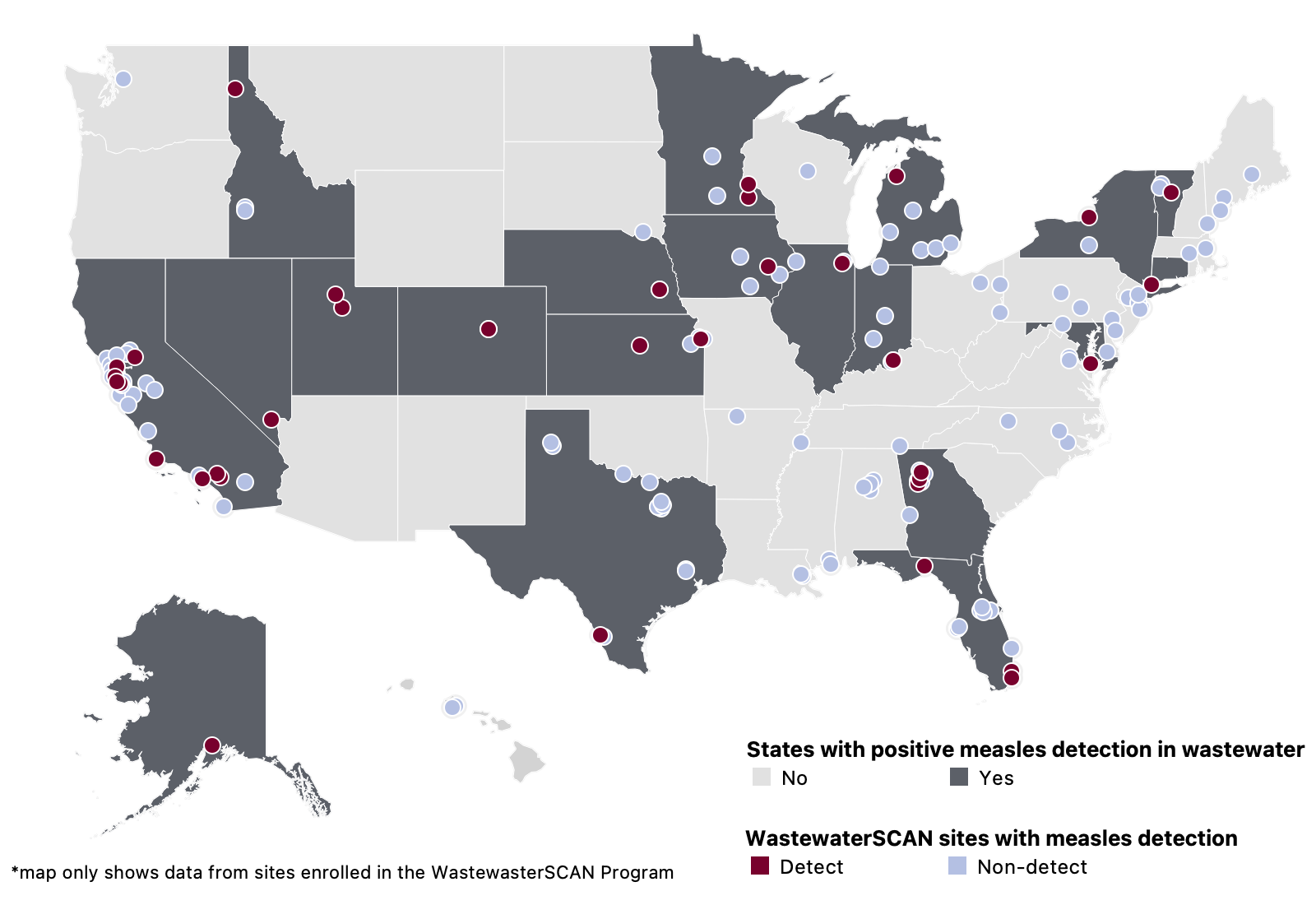
The WWSCAN program began monitoring for measles in wastewater start mid May 2025. This map only shows data from the sites/states that are covered by the WWSCAN program.
Below are the current MeV wastewater categories for all WWSCAN sites (the category is determined by the frequency of detection).
- 138 are in the NOT-DETECTED Wastewater Category
- 0 are in the LOW Wastewater Category
- 1 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
Mpox clade Ib DNA - In mid December 2024, WWSCAN began prospective monitoring for Mpox clade Ib. You can find a heat map of the Mpox clade Ib data here. In February 2026, there were no positive detections of Mpox clade Ib. Nationally, Mpox clade Ib is in the 'NOT-DETECTED' category due to no recent detections.
Below are the Mpox clade Ib wastewater categories for all WWSCAN sites (recall the category is determined by the frequency of detection).
- 139 are in the NOT-DETECTED Wastewater Category
- 0 are in the LOW Wastewater Category
- 0 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
Mpox clade II DNA - There were 3 positive detection of Mpox clade II in February 2026 (1 in CA, 1 in GA, and 1 in IN). Nationally, Mpox clade II is in 'NOT-DETECTED' category due to no recent detections in the past 10 days.
Below are the Mpox clade II wastewater categories for all WWSCAN sites (recall the category is determined by the frequency of detection).
- 138 are in the NOT-DETECTED Wastewater Category
- 1 are in the LOW Wastewater Category
- 0 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
West Nile Virus RNA - In mid October 2025, WWSCAN began prospective monitoring for West Nile Virus (WNV). Nationally, WNV in the 'NOT-DETECTED' category due to no recent detections.
Below are the WNV wastewater categories for all WWSCAN sites (recall the category is determined by the frequency of detection).
- 139 are in the NOT-DETECTED Wastewater Category
- 0 are in the LOW Wastewater Category
- 0 are in the MEDIUM Wastewater Category
- 0 are in the HIGH Wastewater Category
- 8 do not have enough data to calculate
WWSCAN protocols and methods
Protocols (Pre-analytical methods and SARS-CoV-2 analytical methods including controls):
- High Throughput RNA Extraction and PCR Inhibitor Removal of Settled Solids for Wastewater Surveillance of SARS-CoV-2 RNA
- High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses
- Quantification of SARS-CoV-2 variant mutations (HV69-70, E484K/N501Y, del156-157/R158G, del143-145, LPPA24S, S:477-505, and ORF1a Del 141-143) in settled solids using digital RT-PCR
- High Throughput SARS-COV-2, PMMoV, and BCoV quantification in settled solids using digital RT-PCR
- High Throughput wild type measles quantification in settled solids using digital RT-PCR
Pre-prints and peer-reviewed publications provided WWSCAN methods:
- SARS-CoV-2
- Influenza, Human metapneumovirus, Respiratory syncytial virus, Human parainfluenza
- Mpox clade Ib
- Mpox clade II
- Norovirus GII
- Adenovirus group F, Rotavirus
- Candida auris
- Hepatitis A
- Enterovirus D68
- Parovirus
Join us at the next WWSCAN Stakeholder call on March 13, 2026
If you have questions about the website or anything else, please email: wwscan_stanford_emory@lists.stanford.edu
