Bay Area SCAN weekly update 5/31/23
This blog post describes data collected at 8 wastewater treatment plants in the Greater Bay Area of San Francisco, CA, including Sacramento, that are partners in the SCAN project which began in late 2020.
All samples picked up by the couriers as of 5/29/23 have been processed and their data are on the site: wbe.stanford.edu. The data from the SCAN sites are also on data.wastewaterscan.org. Please email Amanda Bidwell at albidwel@stanford.edu if you identify any bugs on the site.
COVID-19
SARS-CoV-2 and Variants
SARS-CoV-2 N gene concentrations at SCAN sites are similar to what they were two weeks ago at most sites. Concentrations are still approximately 100,000 copies/g, still well above our lower detection limit. An example chart from Sacramento is below.

We ran a 2-week look period trend analysis on the plants for the N gene target using methods outlined in this peer-reviewed paper by Chan et al. from our team (link to paper). Palo Alto has significant downward trend, while the rest of the plants have no trend.
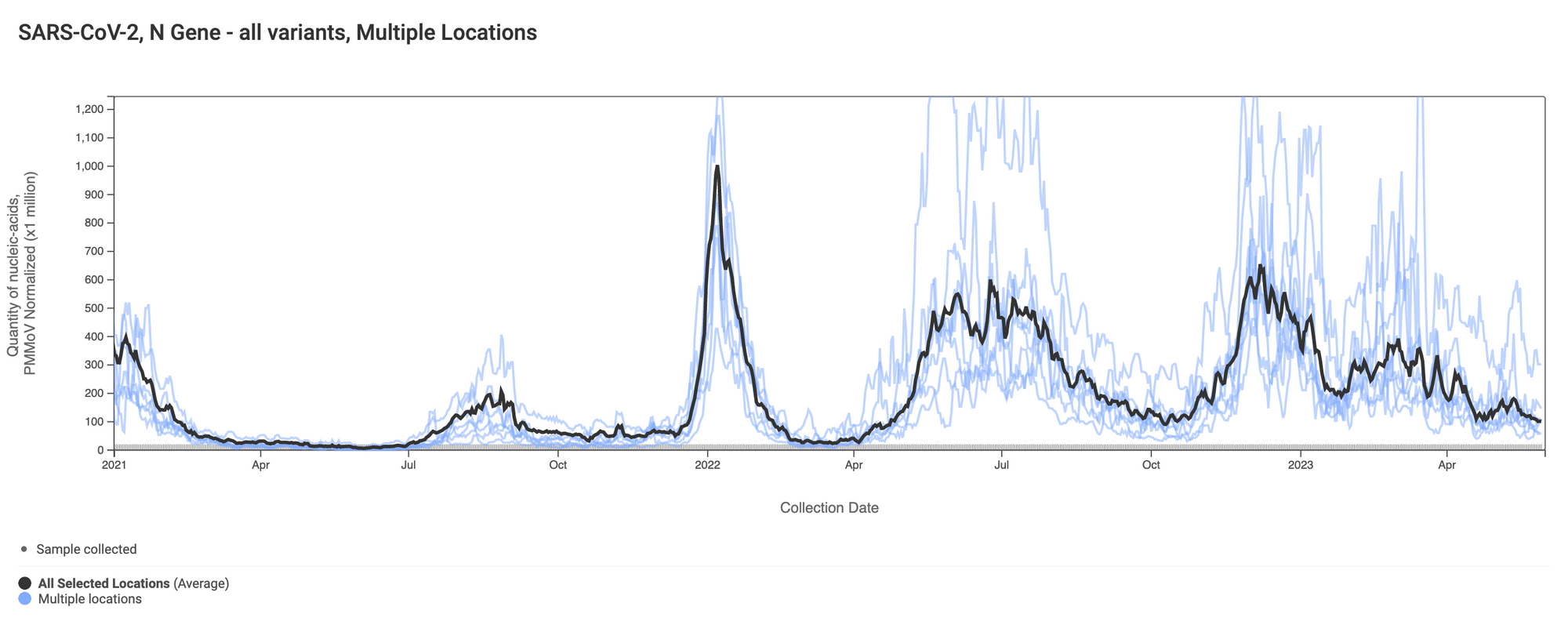
Here are all the data from all the SCAN plants together for the duration of this project. You can access the graph here. The population weighted average across the plants is shown in black.
The chart from Sacramento (2 charts above) shows that the mutation marker for XBB* (shown in violet) is close to the blue line (showing the N gene, present in all variants). This indicates that most of the individuals shedding SARS-CoV-2 RNA in the sewersheds are infected with and shedding XBB* rather than another variant. We see the pattern across all the sewersheds in SCAN. The assay we use for XBB* detects XBB.1.16 as well as XBB.1.9 and FD.2 which are emerging XBB*.
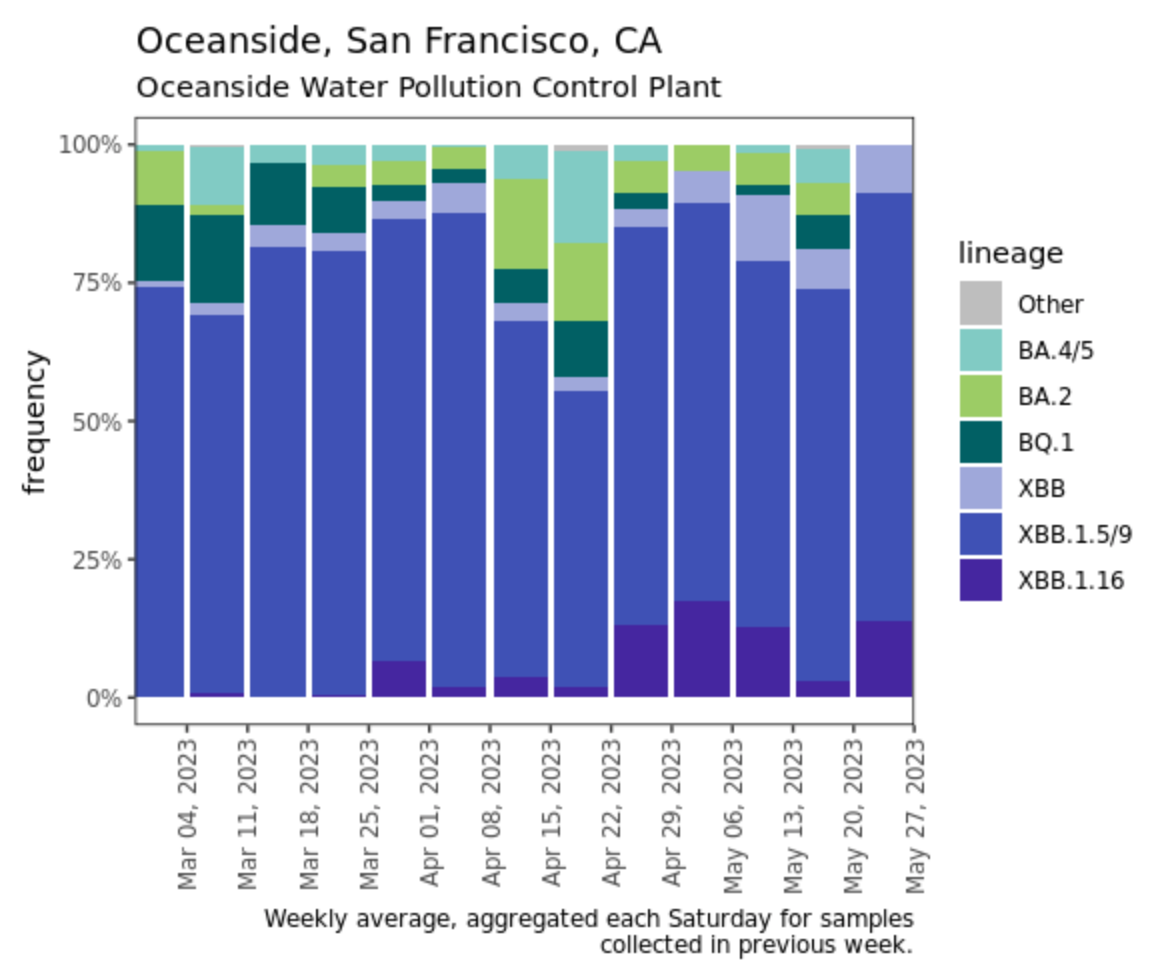
We have detected XBB.1.16 in wastewater from the three SCAN plants we sequence SARS-CoV-2 from (Oceanside, San Jose, and Sacramento). The data from the last batch of samples processed (week of May 20) suggests the sublineage could make up ~10% of the genomes. The chart below shows the fraction of genomes in Oceanside wastewater over time based on sequencing of the S gene. The deep purple shows XBB.1.16.
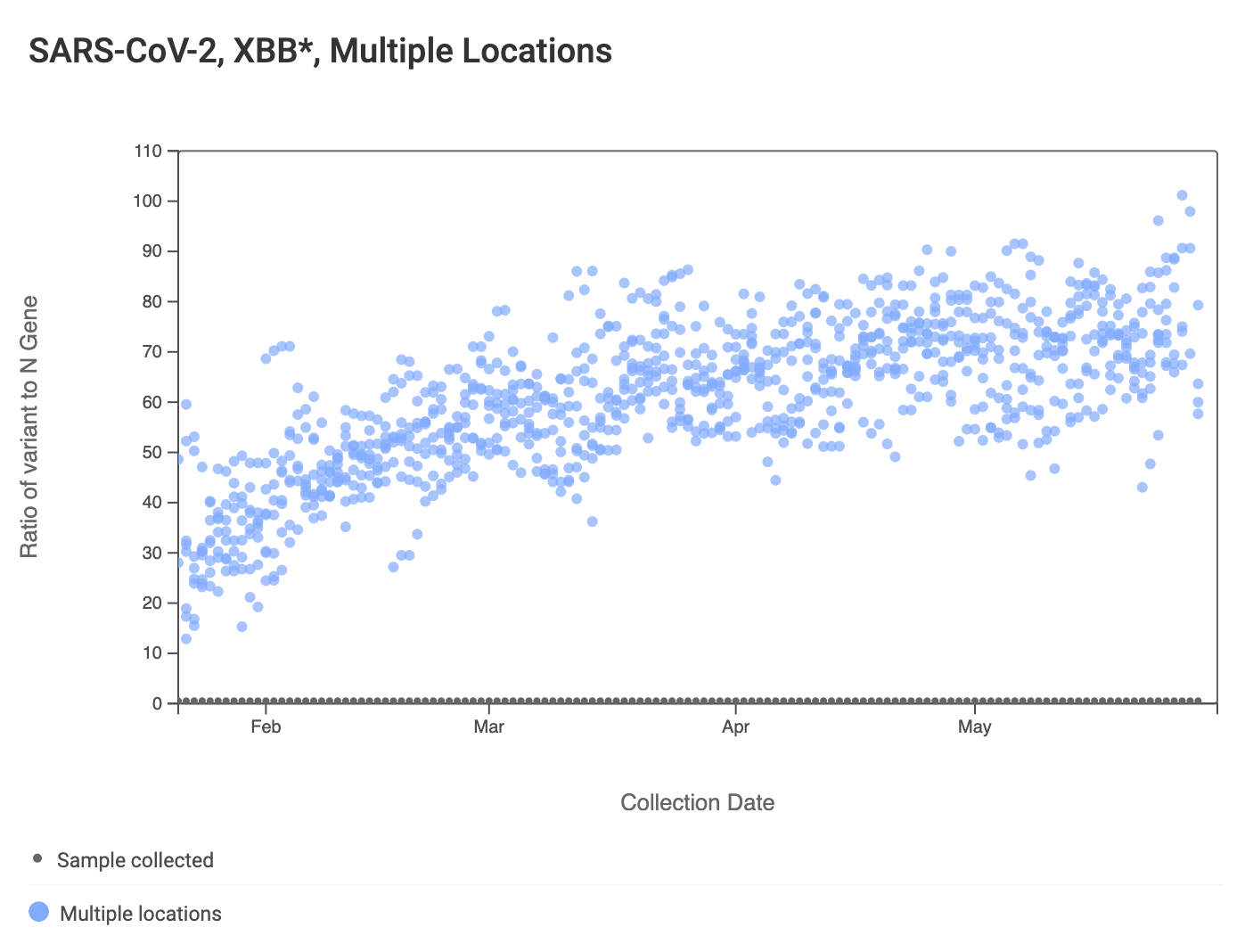
The ratio of the XBB* mutations/N across SCAN plants is shown in the chart below. This chart comes from data.wastewaterscan.org (link here to the chart). As the ratio reaches 100%, it suggests that all the SARS-CoV-2 genomes in wastewater have the XBB* mutations. These data indicate that the regional average ratio XBB* mutations/N is ~80%. These are about the same as last week. These data suggest that the majority of infections across the SCAN communities are caused by XBB*.
Other Respiratory Viruses
IAV, IBV, RSV, and HMPV
Influenza A (IAV) RNA concentrations are very low across the SCAN plants. Concentrations are generally non-detects in samples from recent weeks. The link to the chart below is here if you would like to interact with it. The population weighted average line across all the SCAN plants is shown in black.
The 2-week look period trend analysis on IAV RNA at these plants indicates no trend at all the SCAN plants.

Influenza B (IBV) RNA is sporadically detected at low concentrations at the SCAN plants. Each plant is represented by a row (y-axis), and dates are shown along the x-axis. Blue indicates a non-detect, and darker orange/red is proportional to concentration with the highest values being darkest. A white means no sample was collected. Here is the link to the heat map chart - the linked chart will update automatically as more data are added to the site.

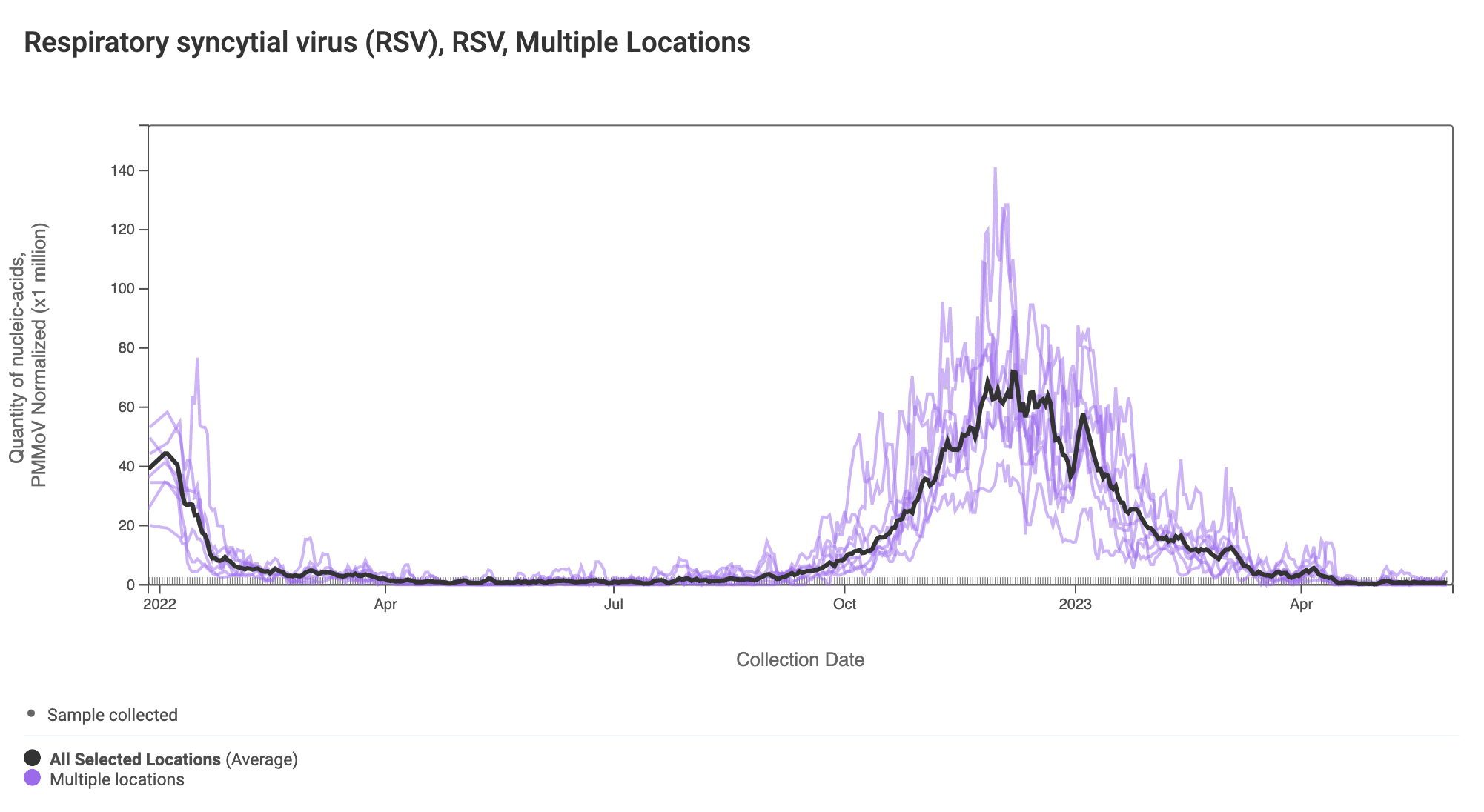
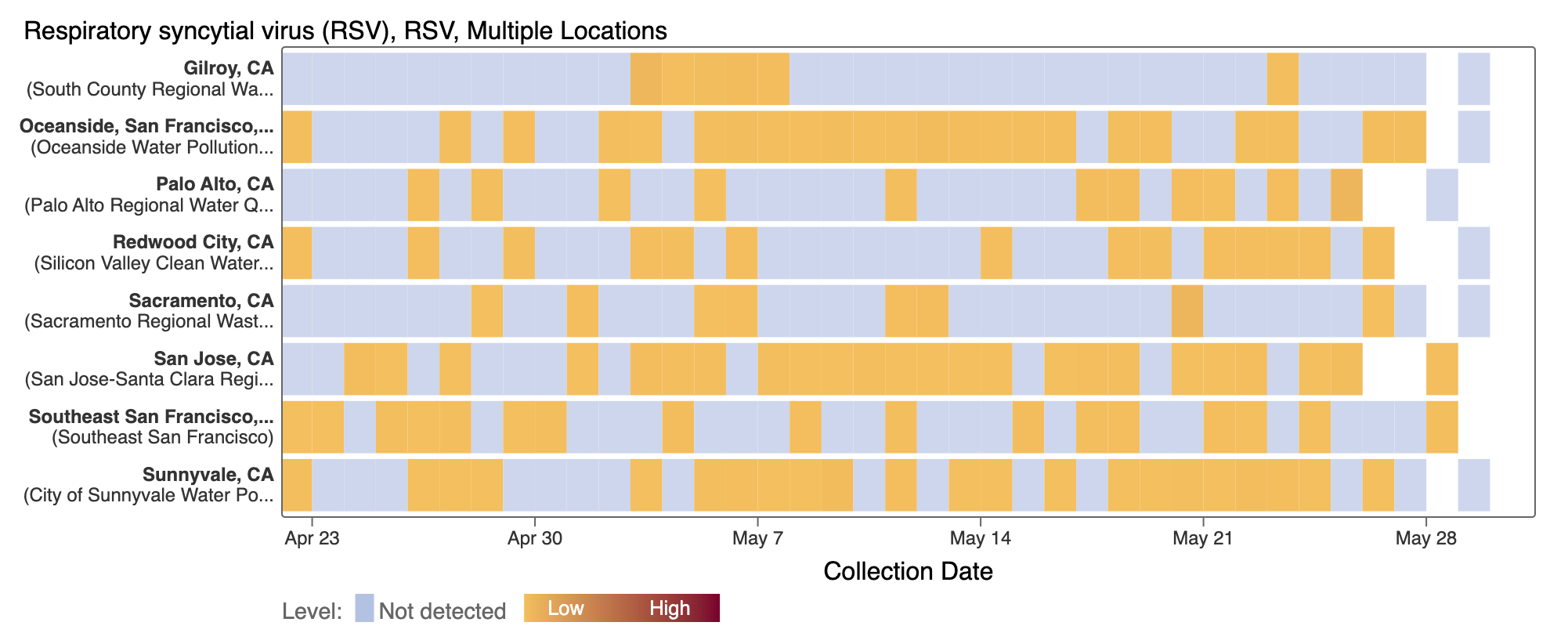
RSV RNA has been non-detectable in most samples in the past week. This plot shows data from all the SCAN sites together since last winter and can be access here. The black line is the population weighted average.
The 2-week look period trend analysis on RSV RNA at these plants indicates no trend at all the plants.
The heat map below shows the recent more frequent occurrence of non-detects for RSV over the last few weeks.
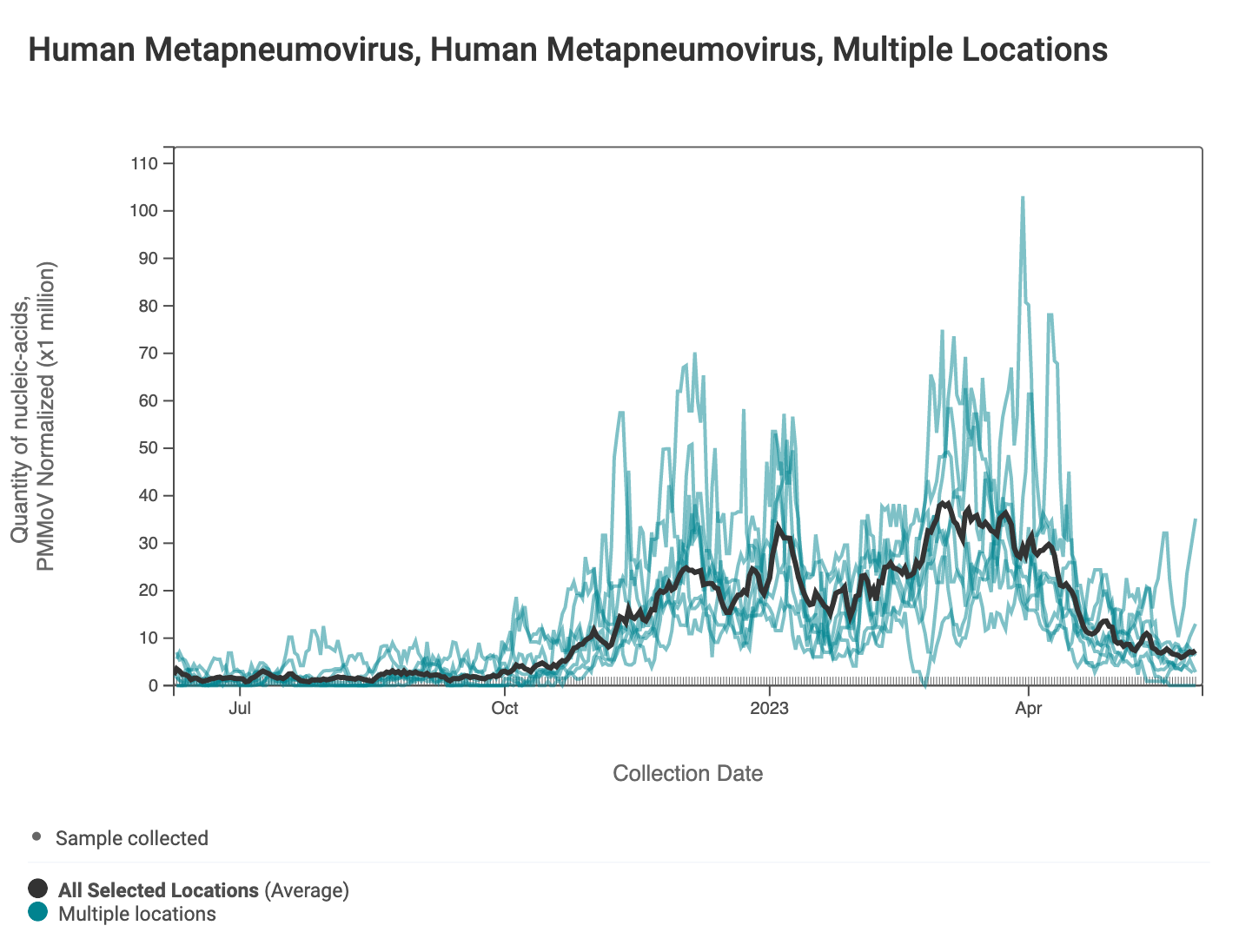
HMPV (human metapneumovirus) RNA concentrations came down across most of plants in SCAN last week, but have been relatively constant overall. Below are charts showing data from all the SCAN plants together (link here to this chart if you want to interact with it).
The 2-week look period trend analysis on HMPV RNA indicates no trend at the plants.
Norovirus
Assay detects human norovirus GII
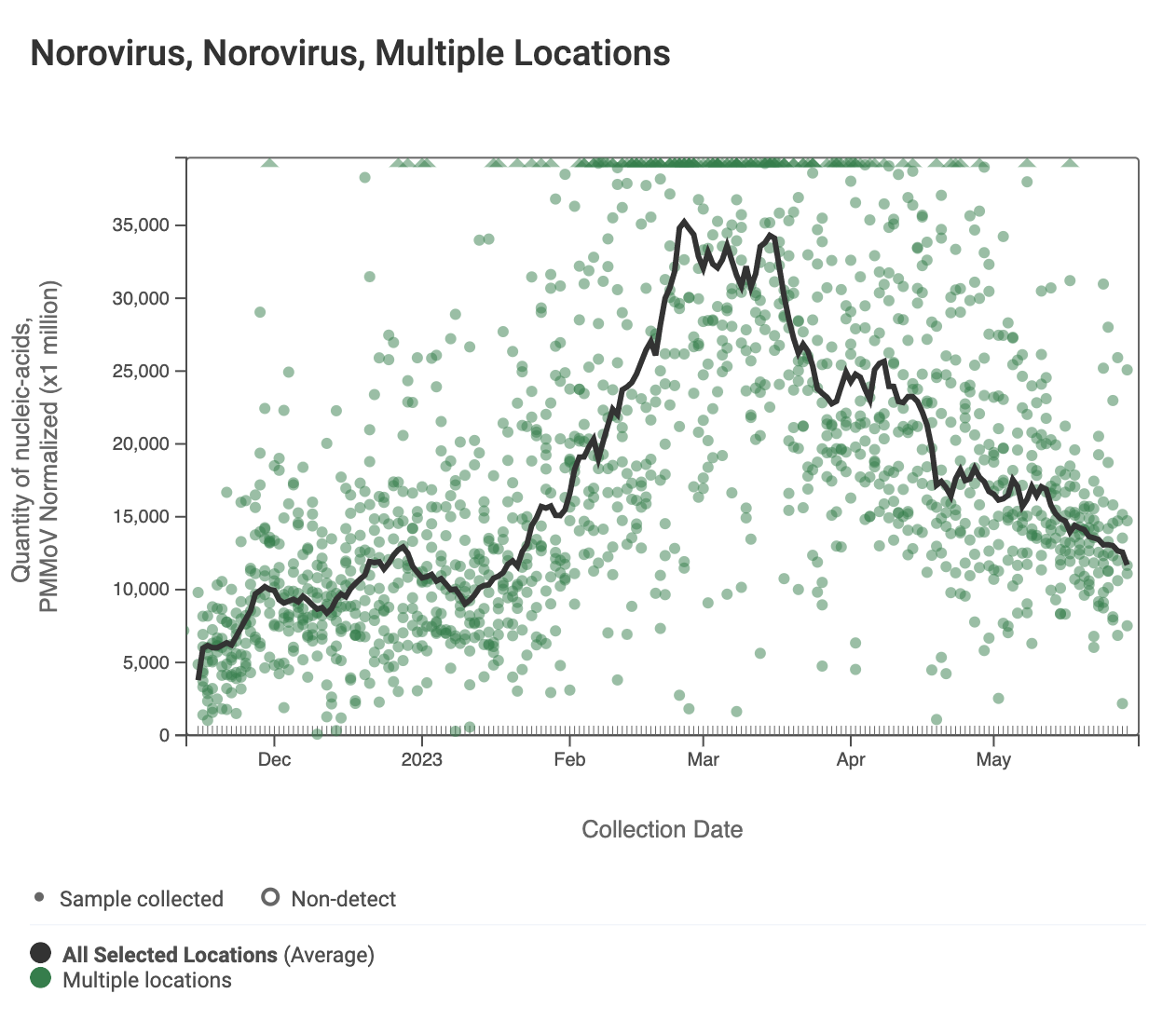
Norovirus GII RNA concentrations are shown in the chart below; all the SCAN plants are shown together. The population weighted average line is shown in black, and that line shows a slow decrease over the last couple weeks. You can interact with the chart of all the plants at this link.
The 2-week look period trend analysis on HuNoV GII RNA these plants indicates a significant downward trend at Oceanside SF, Sacramento, and Sunnyvale, and no trend at the other plants.
Mpox
We've observed non-detects in the last 8 weeks for MPXV DNA at the SCAN plants, with the exception of one sample from 5/7/23 (shown in orange below). The heat map show all the SCAN sites as a row, and each date as a column. The color blue means the sample was non-detect for MPXV DNA and the colors get darker with higher concentrations. White indicates no sample was collected. The charts shows all the data we have collected, almost a year's worth of data. You can access the chart here.
There was recently a national warning that we might see a resurgence in MPOX cases. The data from wastewater suggests that is not the case presently at the SCAN plants.

