Georgia WWSCAN Biweekly Newsletter Update 7/19/24
Welcome to the bi-weekly update for WWSCAN partners in Georgia! The samples provided up through 7/15/24 have been processed in the lab and data are on the site at data.wastewaterscan.org.
If you notice any bugs on the site or have any comments about it, please continue to send your feedback via email wwscan_stanford_emory@lists.stanford.edu. We thank you for your partnership!
For site level summaries of the GA plants with current Wastewater Categories and concentration plots, see the following links:
- Big Creek, Roswell, GA
- College Park, GA
- Columbus, GA
- Johns Creek, Roswell, GA
- Little River, Roswell, GA
- RM Clayton, Atlanta, GA
- South River, Atlanta, GA
- Utoy Creek, Atlanta, GA
Infectious Disease Target Review
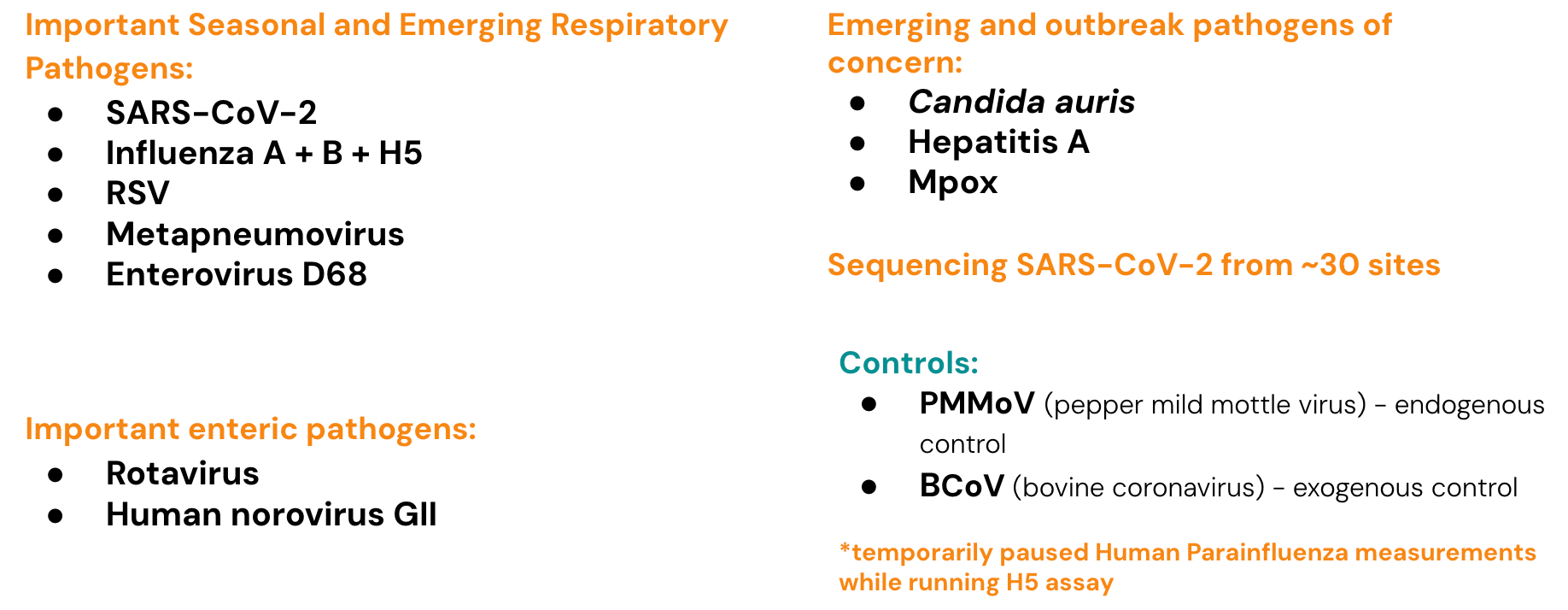
Currently monitoring a suite of Respiratory, Gastrointestinal and Outbreak Pathogens of Concern
The methods for our assays are in the public domain and links for these are provided at the end of the newsletter.
COVID-19
SARS-CoV-2 concentration in wastewater and sequencing for variants
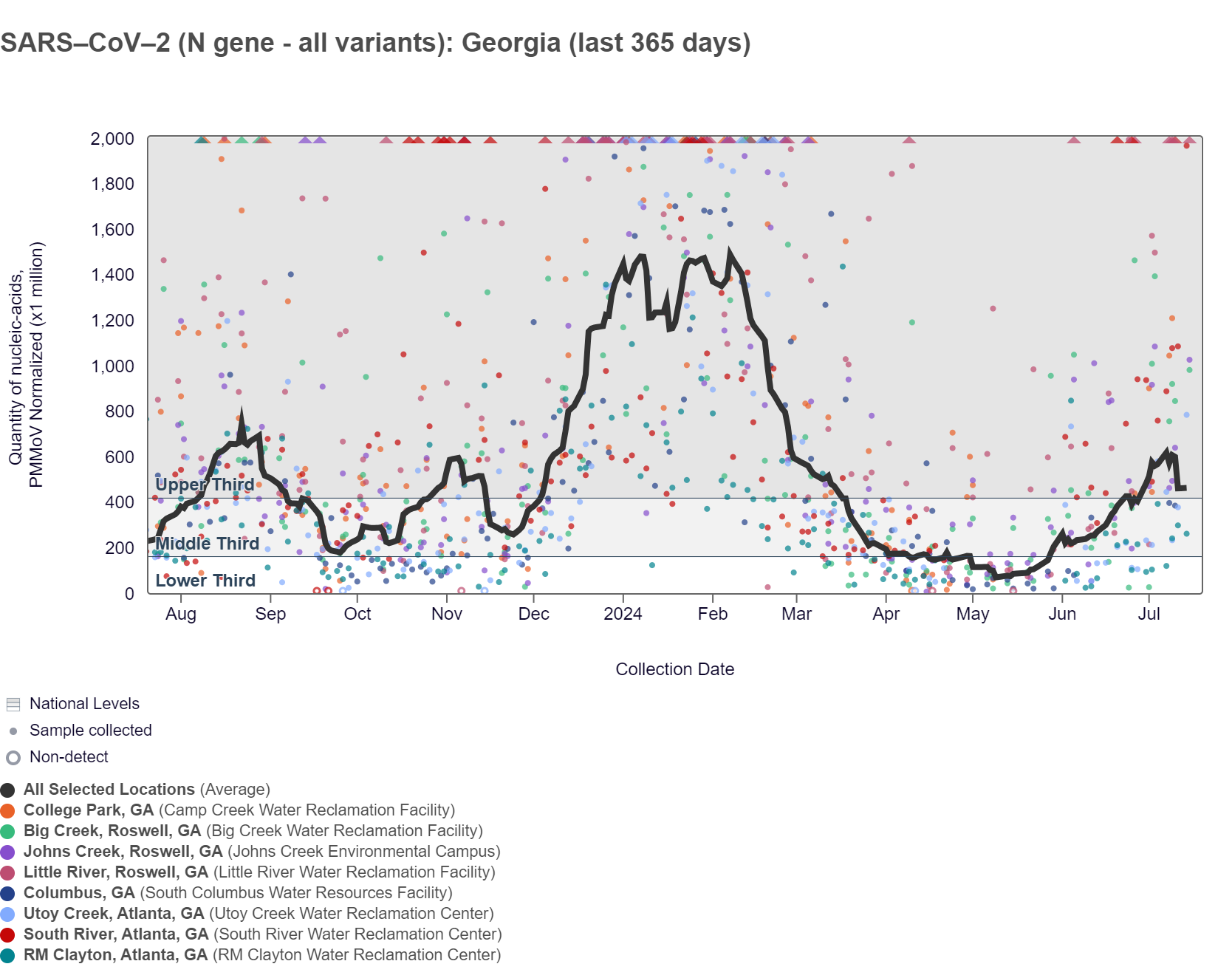
SARS-CoV-2 RNA concentrations have been between 50,000 and 700,000 copies/gram in the last two weeks. The chart below shows the raw data over the last 365 days and the population-weighted aggregated trend line for all 8 Georgia sites when the data is normalized by PMMoV. The aggregated line and the National Levels benchmarks illustrate that SARS-CoV-2 N gene RNA concentrations among Georgia sites are now within the upper third level of all concentrations measured in the last year. SARS-CoV-2 is in the "High" wastewater category nationally and in the Southern region.
As of 7/19/24, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the HIGH (5 sites) wastewater category while RM Clayton is in the MEDIUM (1 site) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
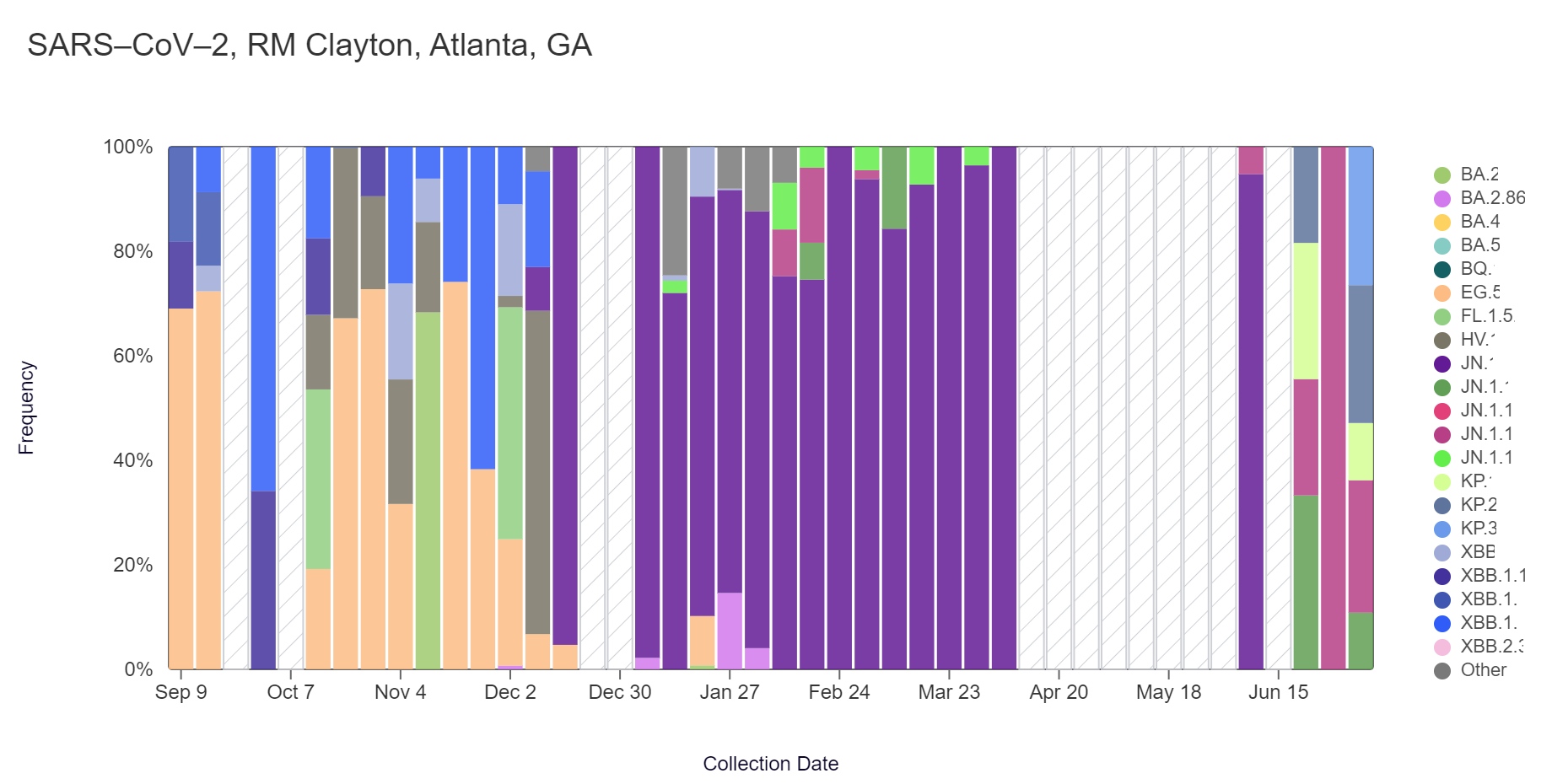
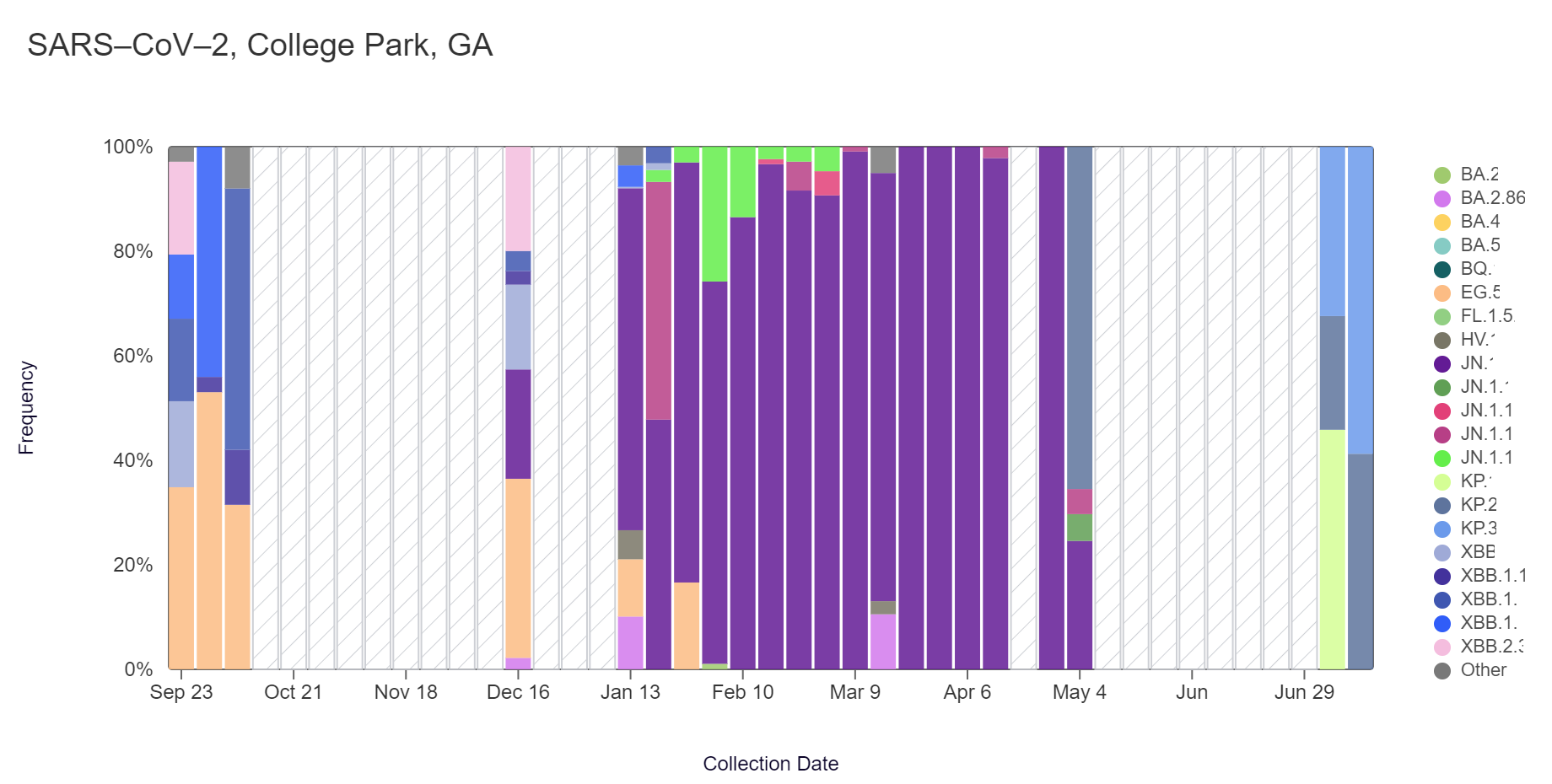
Sequencing of SARS-CoV-2 from wastewater is available for RM Clayton and College Park (Camp Creek). Note that there is a gap in data for RM Clayton and College Park due to a pause in sample collection, which has now resumed! The plots below show the relative proportions of different variants inferred from sequencing the entire genome of SARS-CoV-2 (Note: the sequencing variant plots are now available on the website, so the plots that are shown below are linked above if you want to interact with them). Results are based on sequencing of 2 samples per week, combined to provide a weekly value.
In the most recent samples, KP.3 (26.5%) and KP.2 (26.4%) made up the largest proportions of lineages detected at RM Clayton. KP.3 (58.8%) also made up the largest proportion of lineages detected at College Park. Note that the most recent data is shown below & should be updated as more samples are analyzed (through July 6 at RM Clayton and July 13 at College Park).
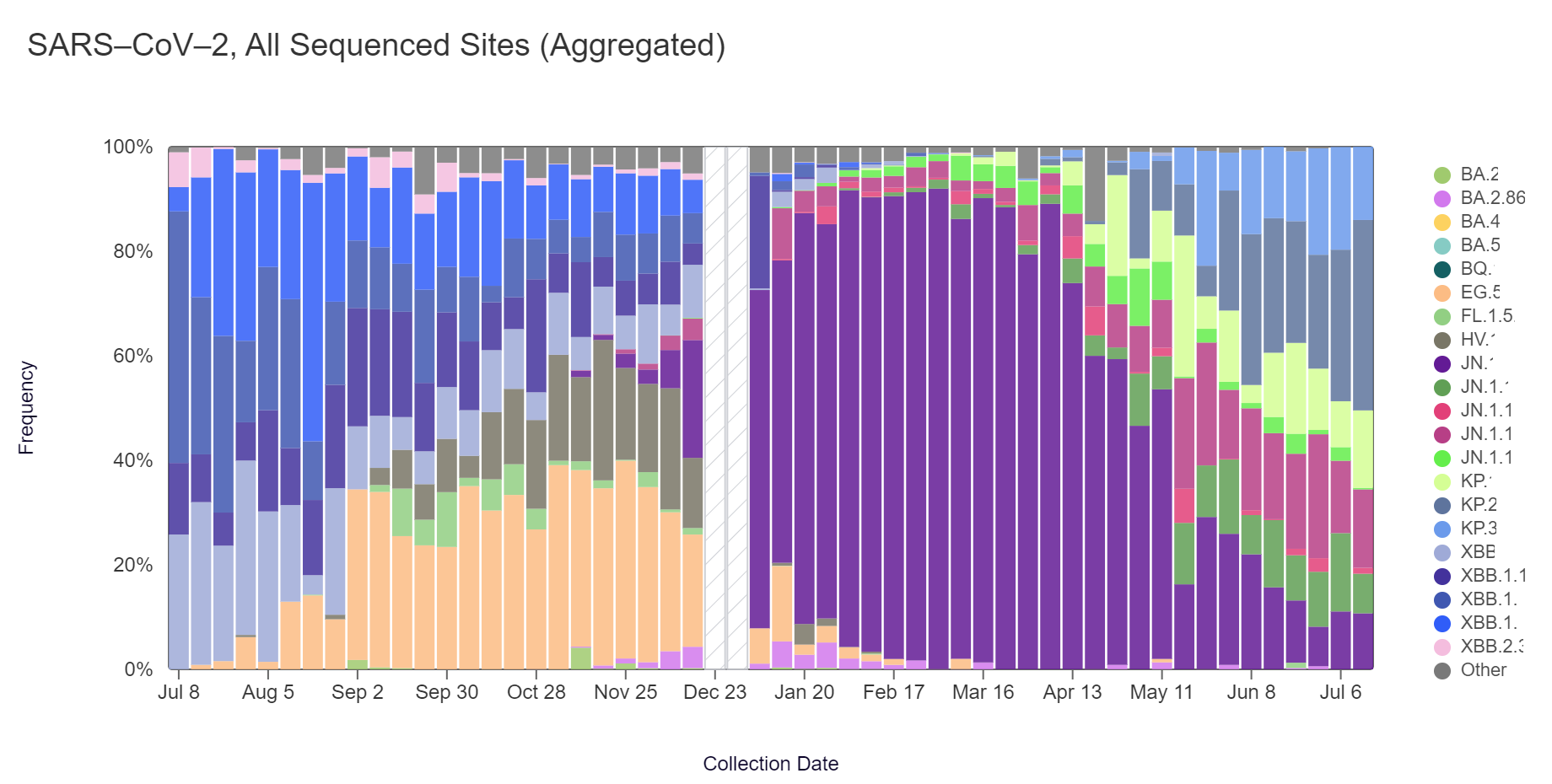
Sequencing data is now also available in aggregate across all sites in WWSCAN with sequencing data through July 13. That plot is shown below and also suggests that KP.2 is the most abundant variant across all sites (36.4%).
Other Respiratory Pathogens
Influenza A and B, Respiratory Syncytial Virus, Human metapneumovirus & EV-D68
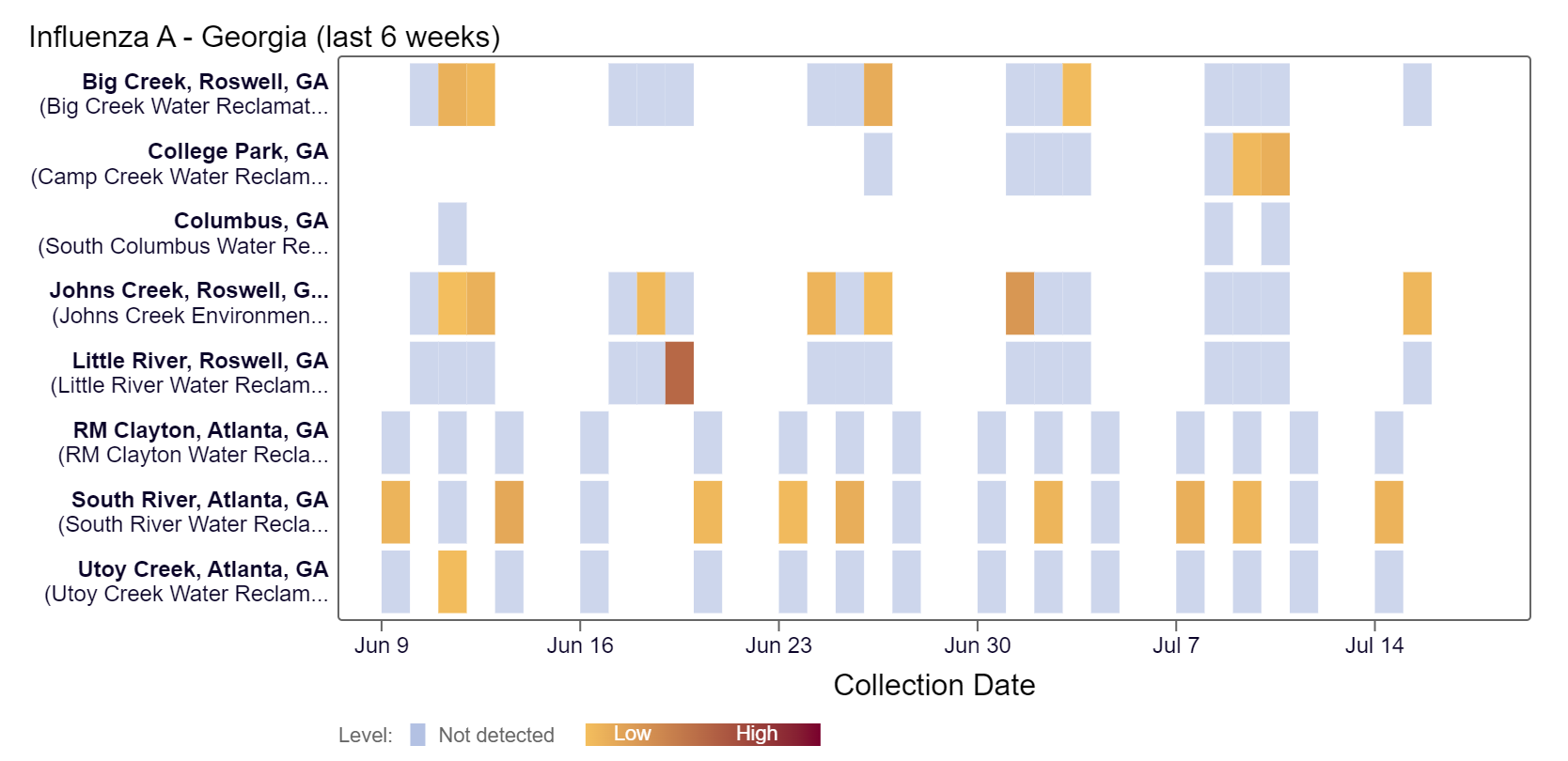
Influenza A (IAV) RNA concentrations continue to be below 5,000 copies/gram in the last two weeks. According to the heat map below, IAV RNA was detected at College Park, Johns Creek, and South River in the last couple of weeks.
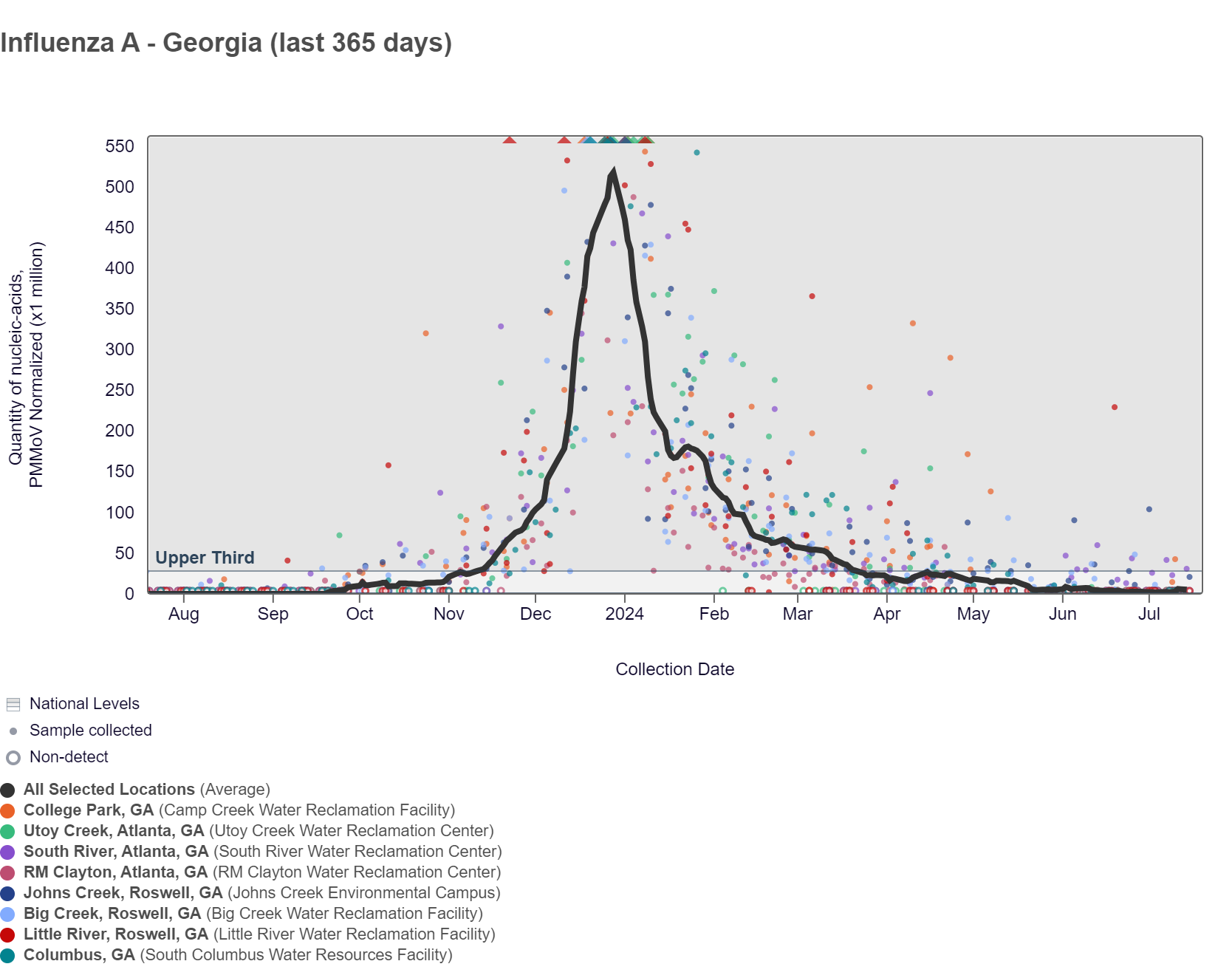
The chart below shows the raw data and the population-weighted aggregated trend line for all 8 Georgia sites when the data is normalized by PMMoV is at the bottom of the middle third level. As of 7/19/24, South River is in the MEDIUM (1 site) wastewater category. RM Clayton, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
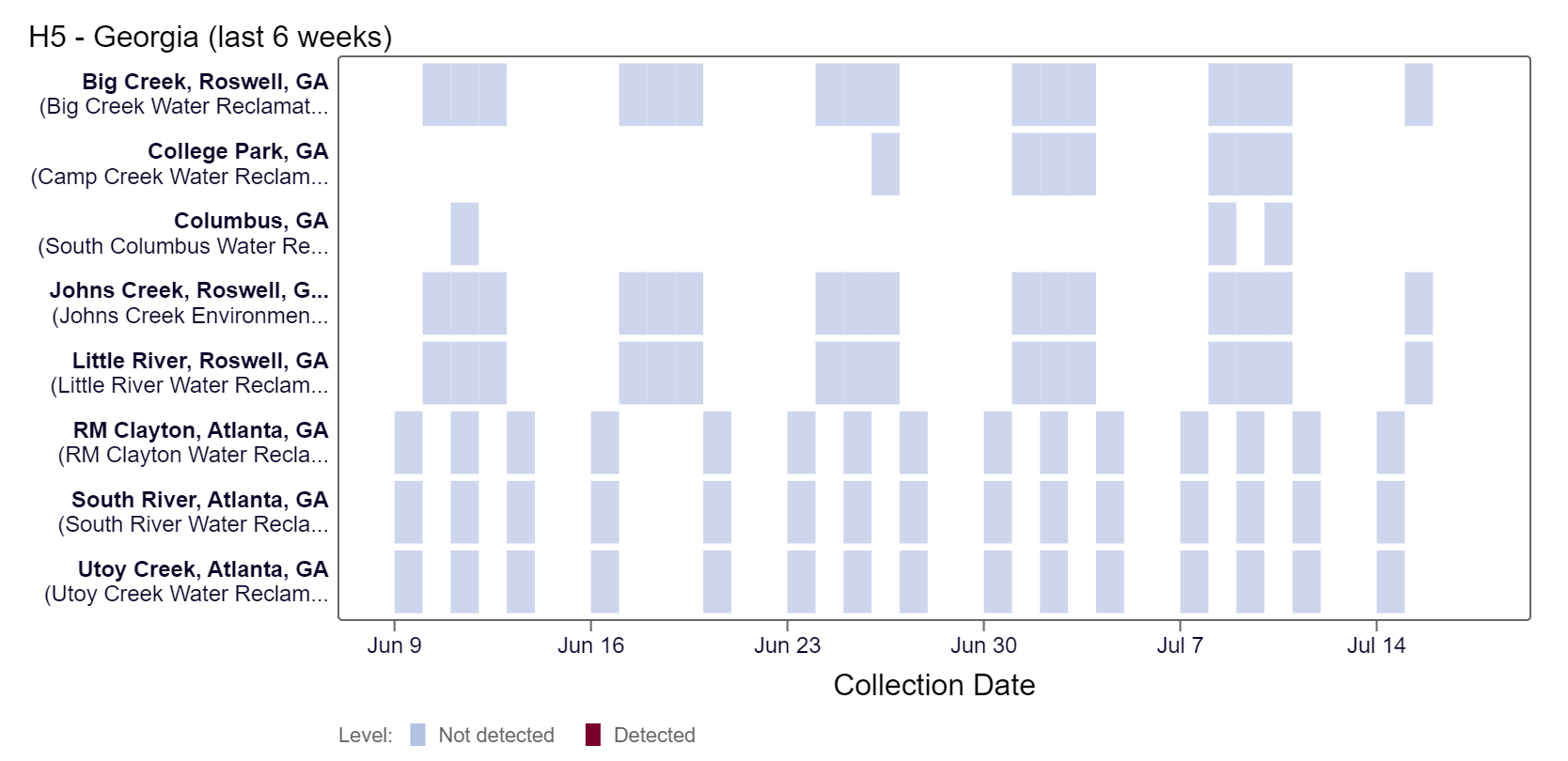
H5 marker in Influenza A (IAV) Below is a heat map showing the Georgia sites with samples collected and tested thus far. Over the last couple of weeks, results have been non-detect at all Georgia sites based on available data. For more information about this testing and interpretation of results, see our FAQ sheet here.
Influenza B (IBV) RNA was detected at the Columbus site within the last two weeks, according to the heat map below.
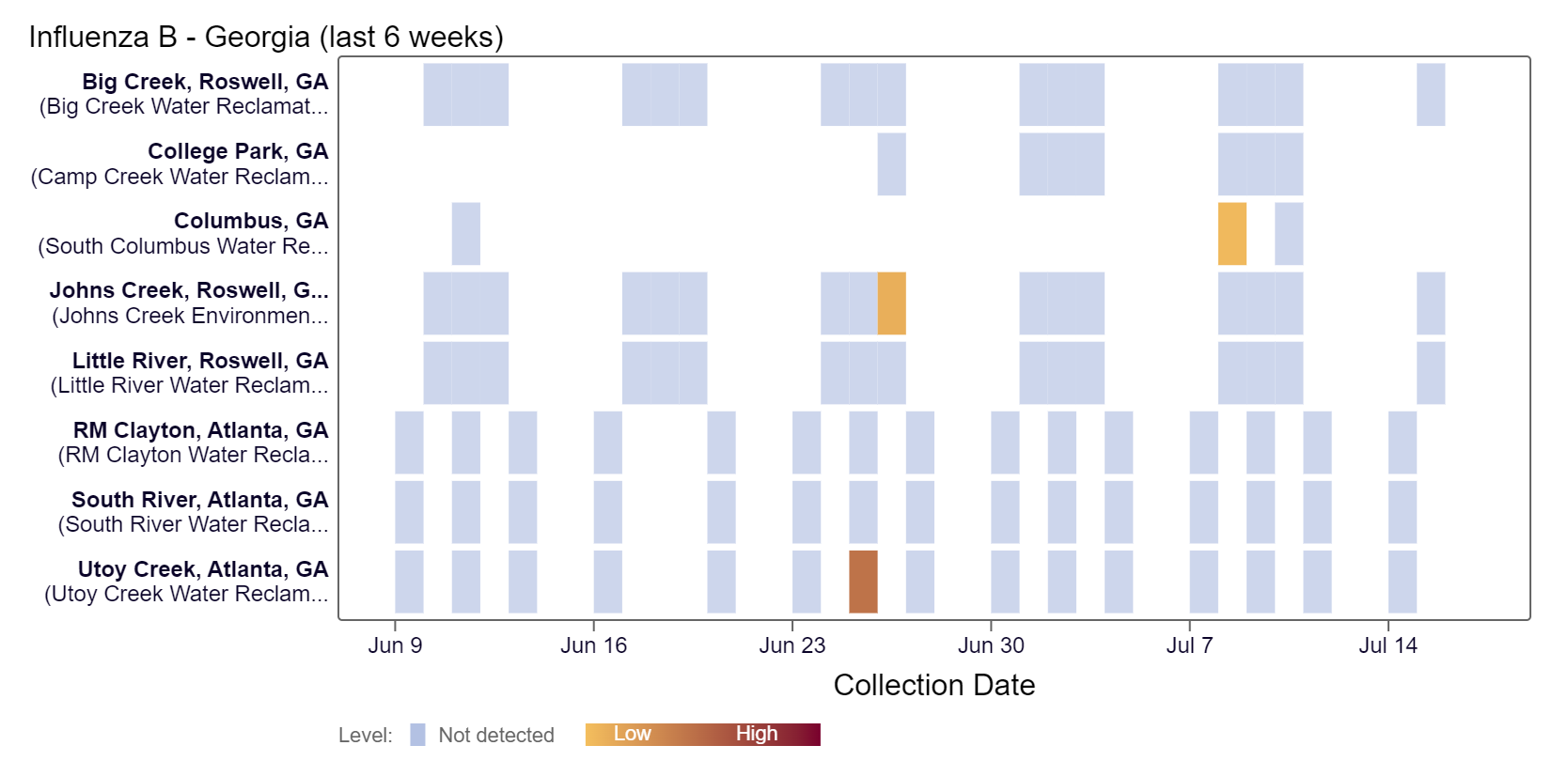
As of 7/19/24, RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
Respiratory syncytial virus (RSV) RNA has been detected at Big Creek over the last two weeks, according to the heat map below.
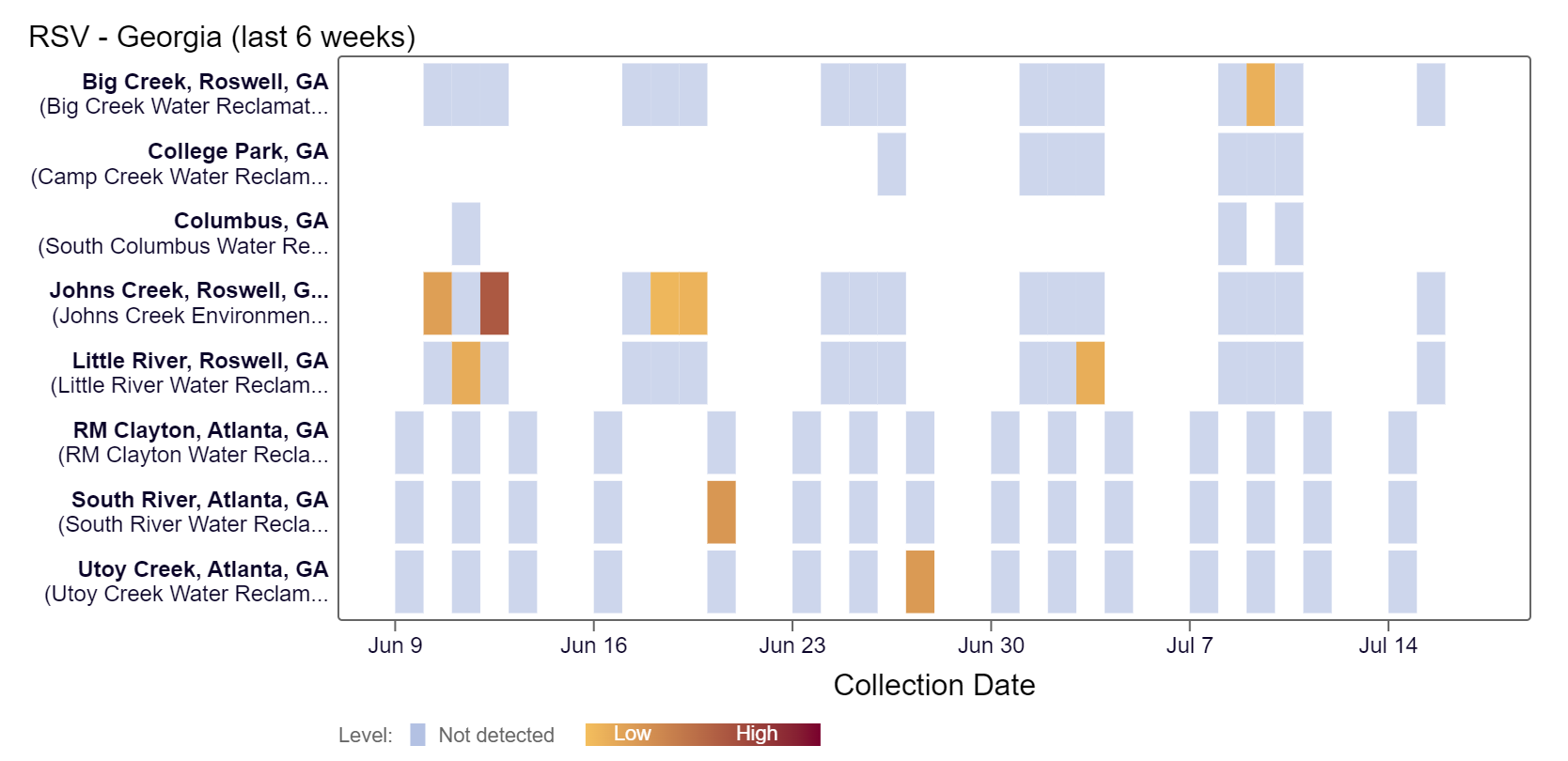
As of 7/19/24, RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
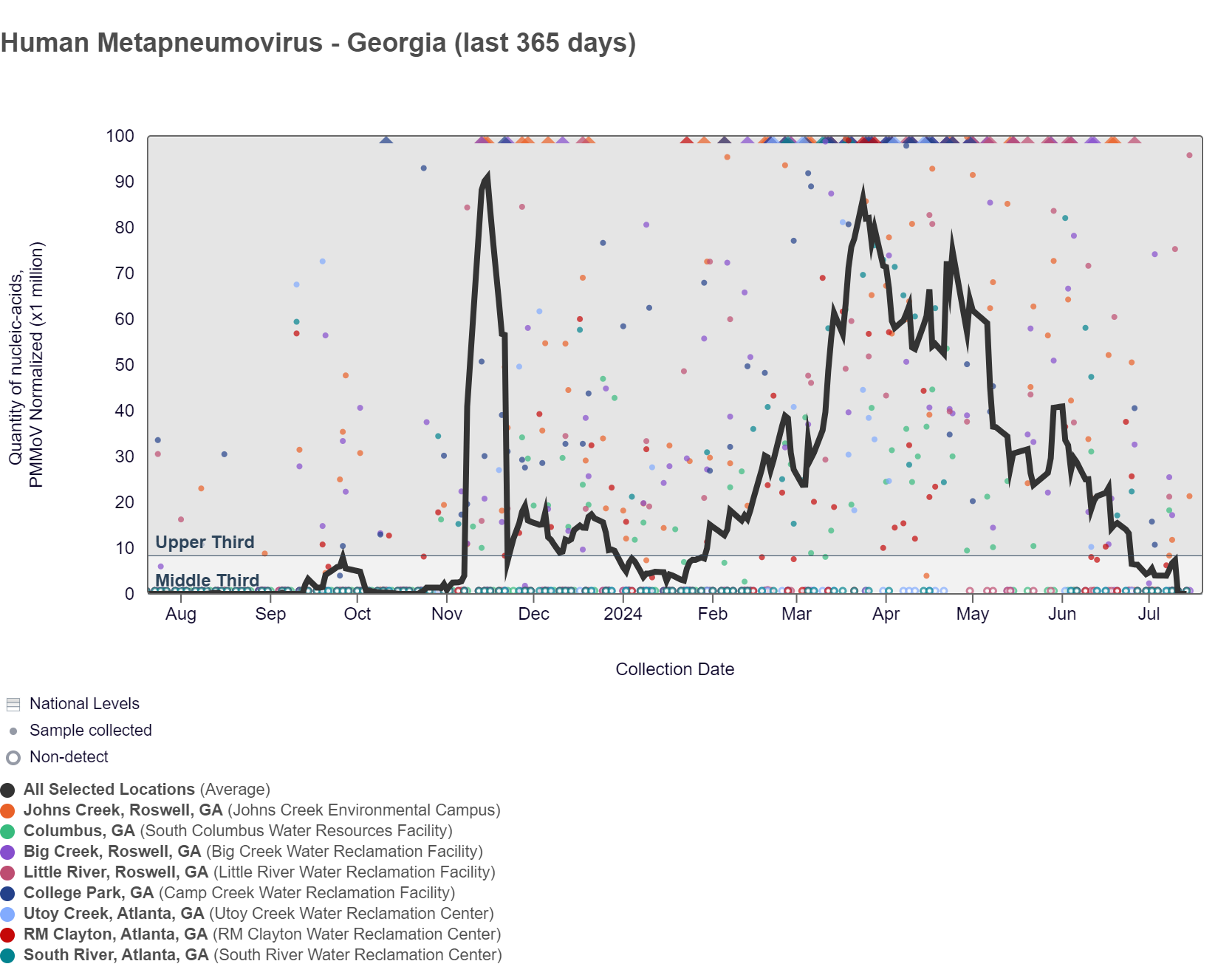
HMPV (human metapneumovirus) RNA concentrations in wastewater among the Georgia sites are below 15,000 copies/g. HMPV RNA concentrations in Georgia are at the bottom of the middle third level of all concentrations measured in the last year. RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category, as of 7/19/24. There was not enough data to calculate a category for Columbus and College Park.
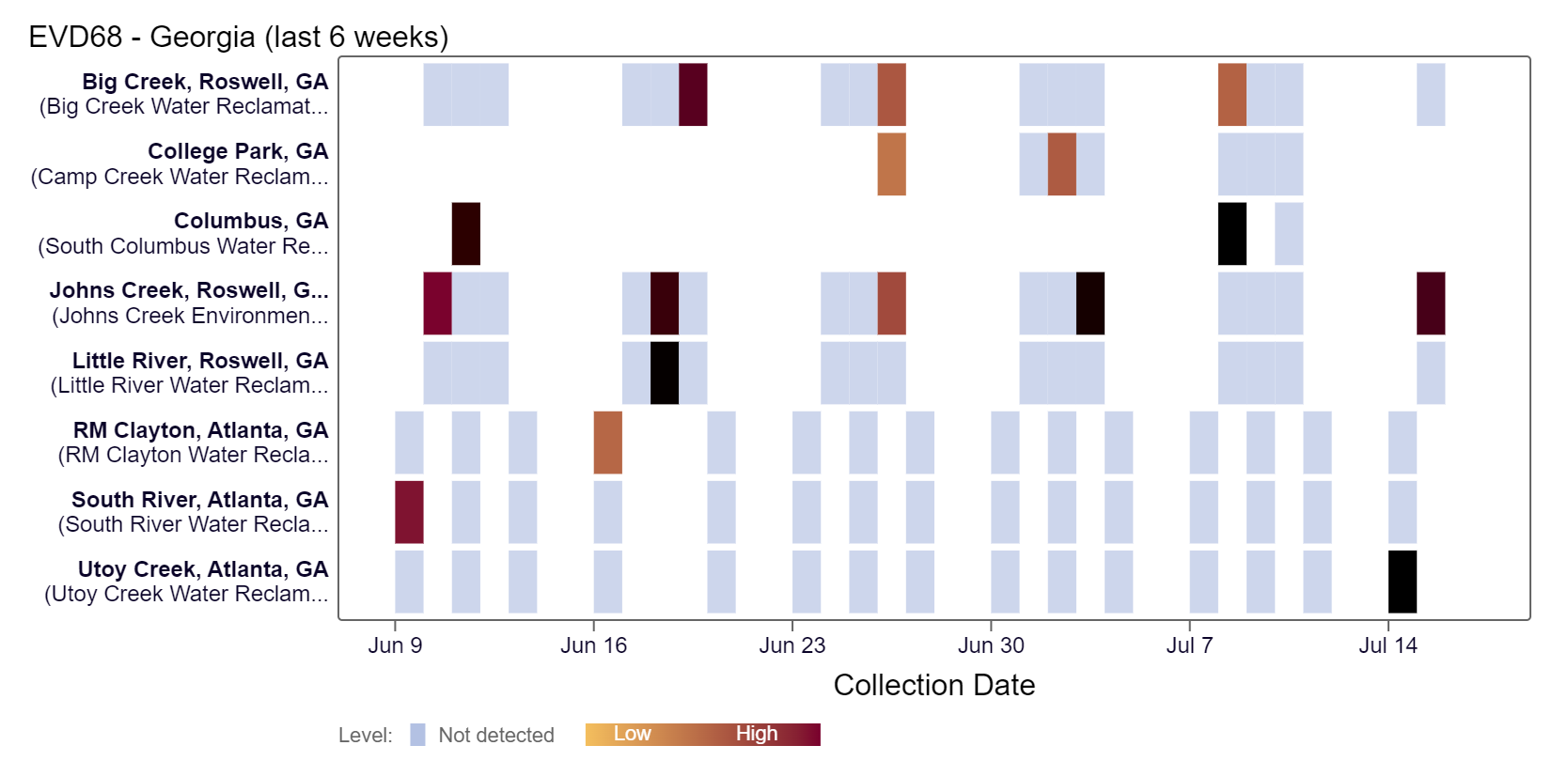
EV-D68 RNA is below 11,500 copies/g with detections at Big Creek, Columbus, Johns Creek, and Utoy Creek sites at high concentrations over the last couple of weeks, according to the heat map below. As a reminder, the color blue means the sample was non-detect for EV-D68 RNA and the colors get darker orange with higher concentrations. As of 7/19/24, RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
Gastrointestinal Pathogens
Norovirus GII and Rotavirus
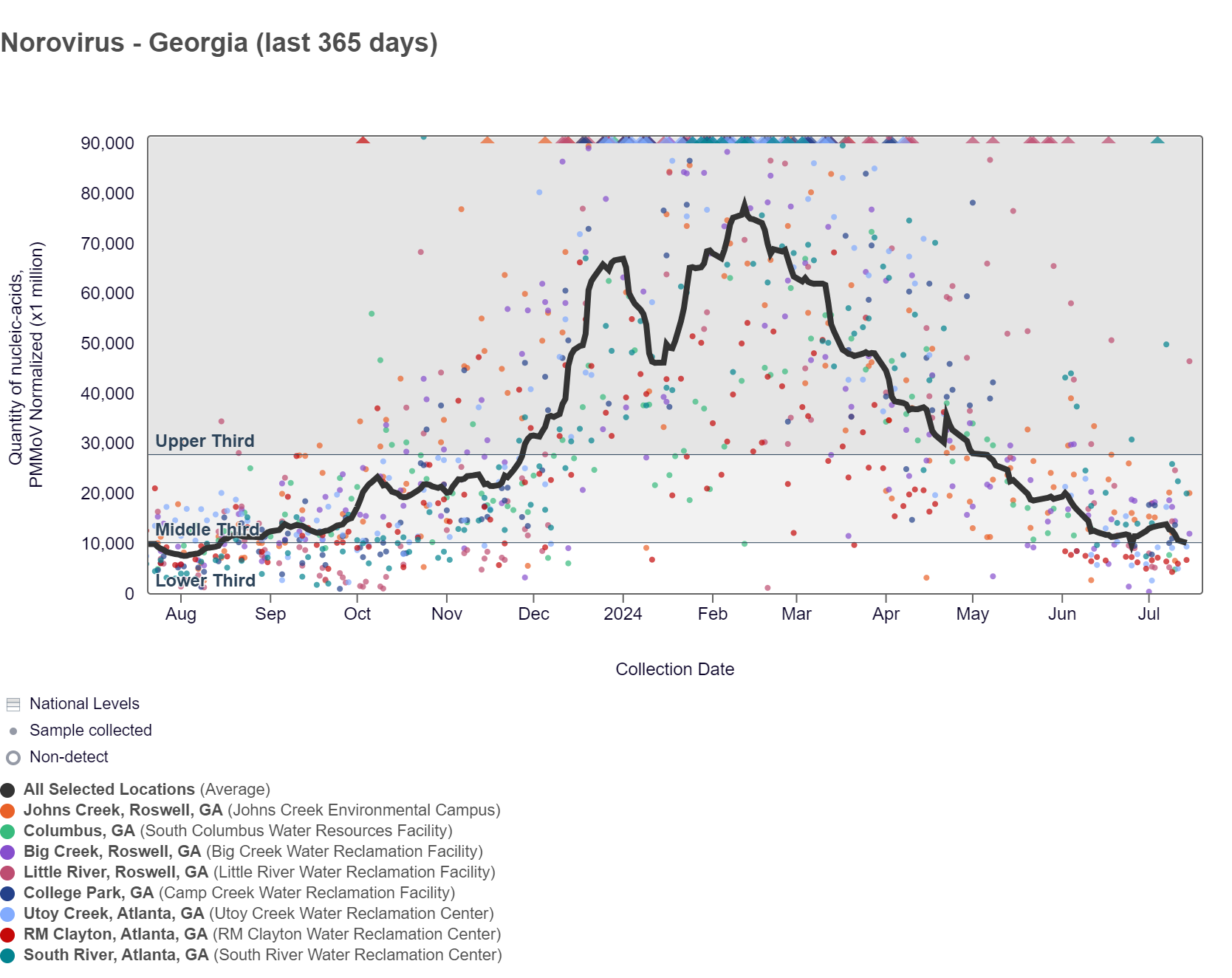
Norovirus GII (HuNoV GII) RNA concentrations range between 750,000 - 7,000,000 copies/g, and the population-weighted average line when the data is normalized by PMMoV for the 8 Georgia sites is between the middle and lower third levels. HuNoV GII RNA chart below also shows the raw data. As of 7/19/24, Little River is in the HIGH (1 site) wastewater category. South River, Big Creek, and Johns Creek are in the MEDIUM (3 sites) wastewater category. RM Clayton and Utoy Creek are in the LOW (2 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
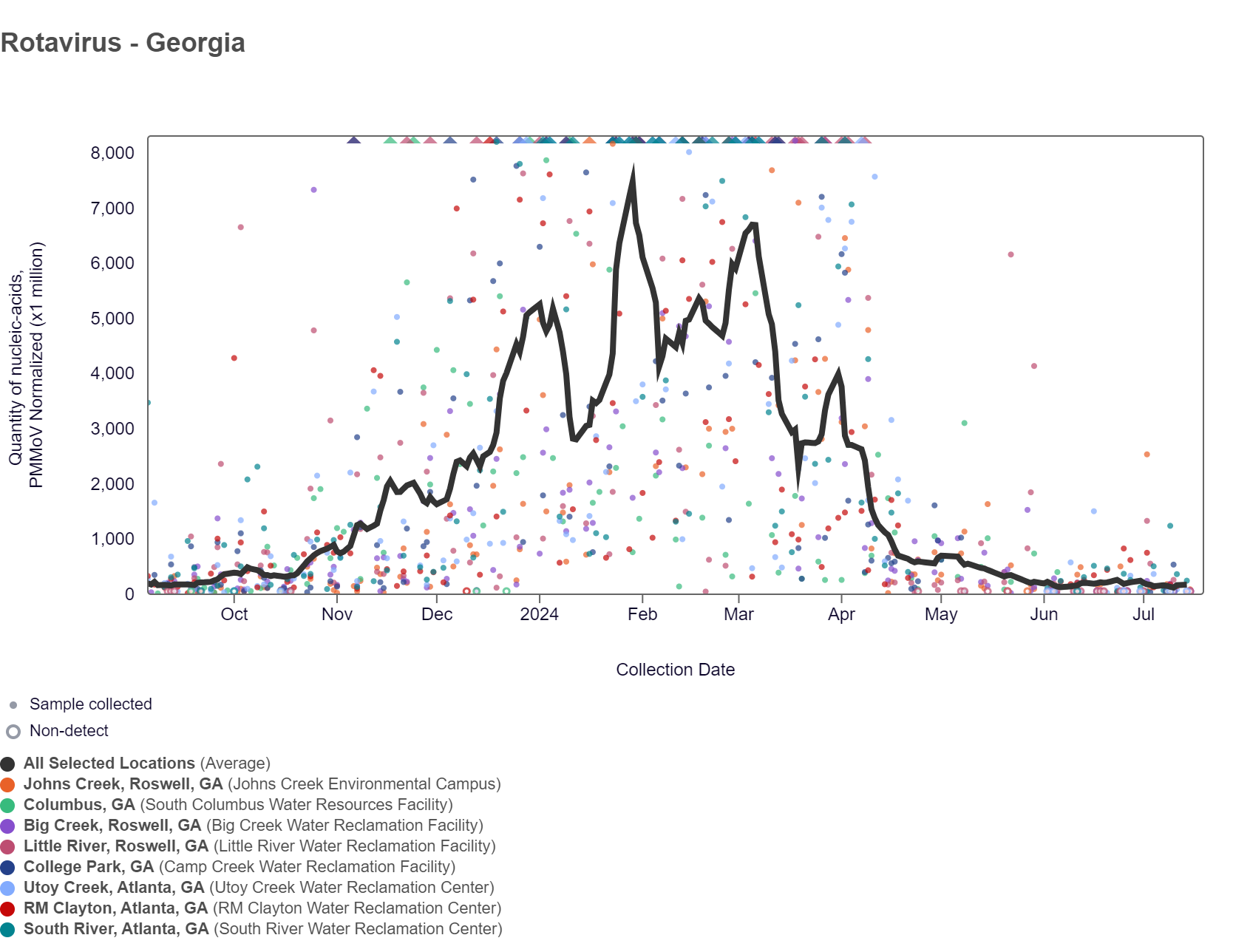
Rotavirus RNA concentrations have been below 100,000 copies/g over the last two weeks. The chart below shows data since September 2023, including the population-weighted average line is shown in black plus the raw data for each site. As of 7/19/24, RM Clayton and South River are in the MEDIUM (2 sites) wastewater category. Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (4 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
Other Pathogens of Concern
Mpox, Candida auris, and Hepatitis A
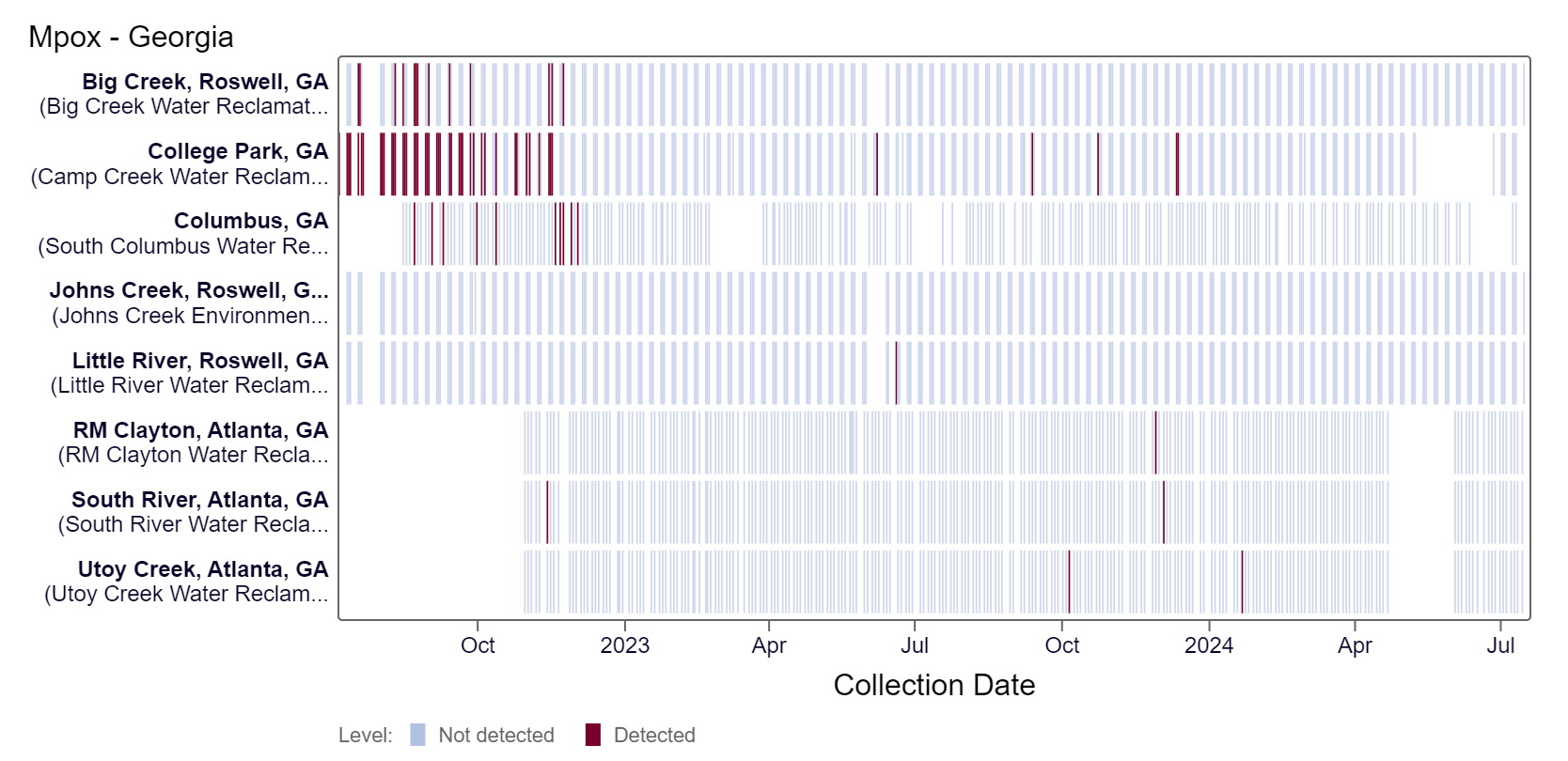
Mpox DNA results have been non-detect at all Georgia sites in the last 21 days. This heat map shows data since July 2022. Sites are labeled in the rows and each date a sample was collected as a column. The color blue means the sample was non-detect for mpox DNA and the color maroon means Mpox DNA was detected. White indicates no sample was collected. RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category as of 7/19/24. There was not enough data to calculate a category for Columbus and College Park.
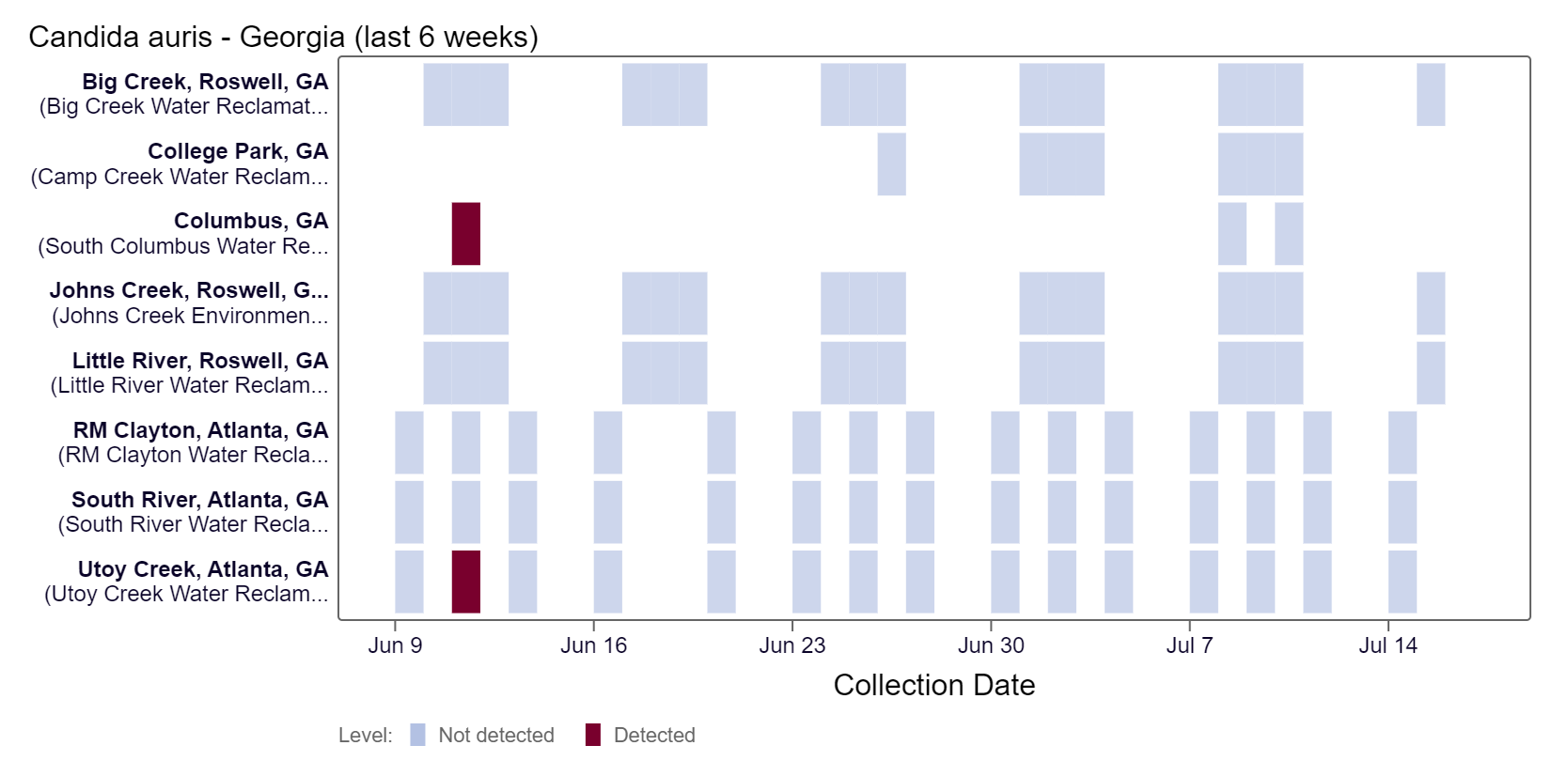
Candida auris DNA has not been detected in the last two weeks, based on the heat map below. As of 7/19/24, RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River are in the LOW (6 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
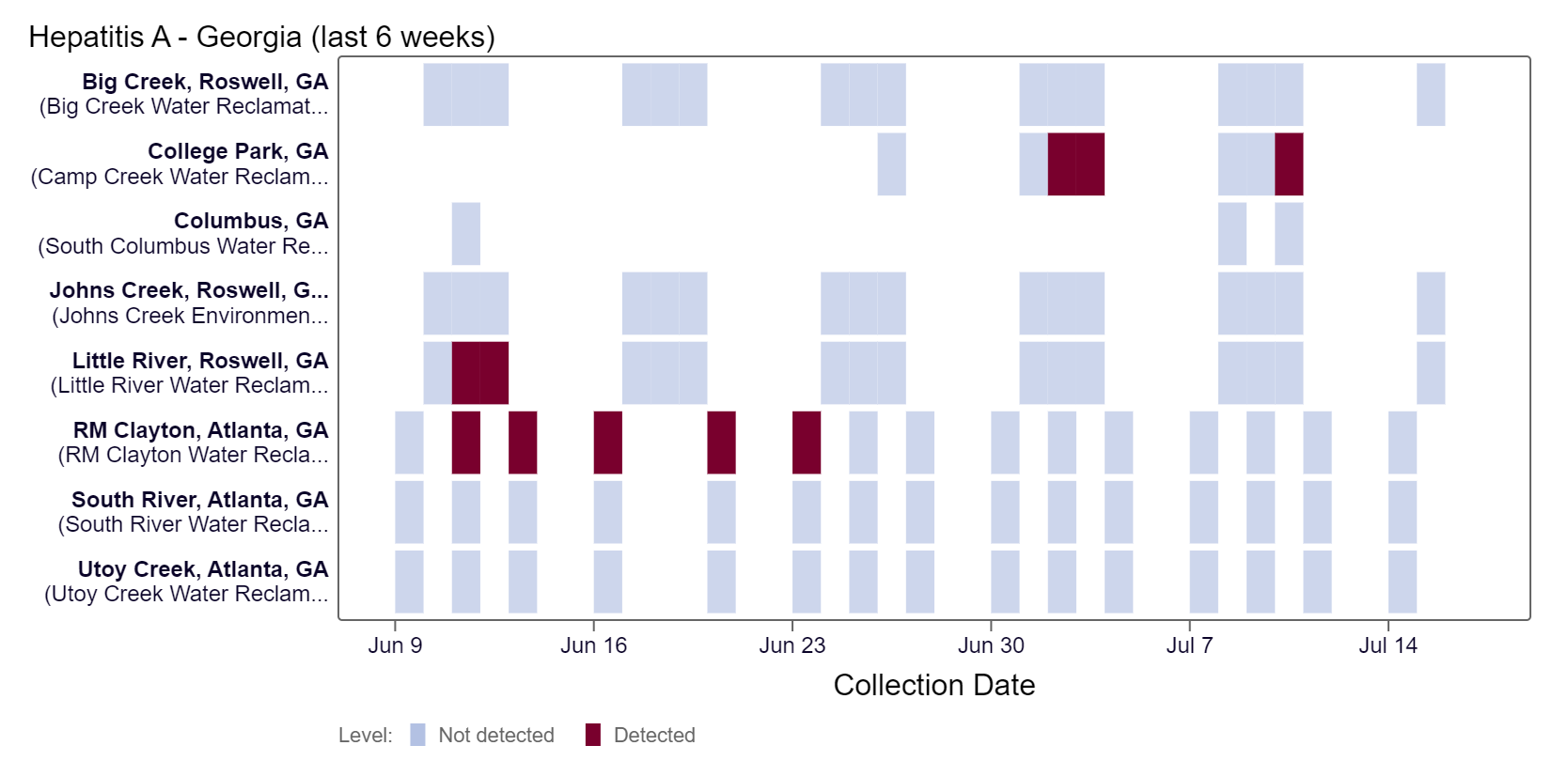
Hepatitis A RNA has been detected at College Park over the last two weeks, according to the heat map. HAV RNA concentrations have been below 4,000 copies/g. As of 7/19/24, RM Clayton, South River, Utoy Creek, Big Creek, Johns Creek, and Little River LOW (6 sites) wastewater category. There was not enough data to calculate a category for Columbus and College Park.
WWSCAN protocols and methods in peer reviewed publications
Protocols (Pre-analytical methods and SARS-CoV-2 analytical methods including controls):
- High Throughput RNA Extraction and PCR Inhibitor Removal of Settled Solids for Wastewater Surveillance of SARS-CoV-2 RNA
- High Throughput pre-analytical processing of wastewater settled solids for SARS-CoV-2 RNA analyses
- Quantification of SARS-CoV-2 variant mutations (HV69-70, E484K/N501Y, del156-157/R158G, del143-145, LPPA24S, S:477-505, and ORF1a Del 141-143) in settled solids using digital RT-PCR
- High Throughput SARS-COV-2, PMMoV, and BCoV quantification in settled solids using digital RT-PCR
Pre-prints and peer-reviewed publications provided WWSCAN methods:
- SARS-CoV-2
- Influenza, Human metapneumovirus, Respiratory syncytial virus, Human parainfluenza
- Mpox
- Norovirus GII
- Adenovirus group F, Rotavirus, Candida auris, Hepatitis A
- Enterovirus D68
Newly published papers are available here
- Spatial and temporal variation in respiratory syncytial virus (RSV) subtype RNA in wastewater and relation to clinical specimens
- Detection of Hemagglutinin H5 Influenza A Virus Sequence in Municipal Wastewater Solids at Wastewater Treatment Plants with Increases in Influenza A in Spring, 2024
- Public health policy impact evaluation: A potential use case for longitudinal monitoring of viruses in wastewater at small geographic scales
Public Health Factsheets:
- Influenza A Fact Sheet: Provides a summary of a November 2023 paper exploring how to analyze wastewater data to extract relevant public health insights on Influenza A. Click here to access the IAV fact sheet.
- RSV Fact Sheet: Provides a summary of a March 2024 paper exploring how to analyze wastewater data to extract relevant public health insights on RSV. Click here to access the RSV fact sheet.
New WastewaterSCAN Published Body of Work document:
- We are excited to share a new document that provides an overview of the program’s peer-reviewed scientific literature, organized by topics and themes. Click here to access the WastewaterSCAN published body of work.
Related News
The next stakeholder meeting will take place on Friday, August 9th @ 12 PM EST. You can use this zoom link to join. Hope to see you there!
