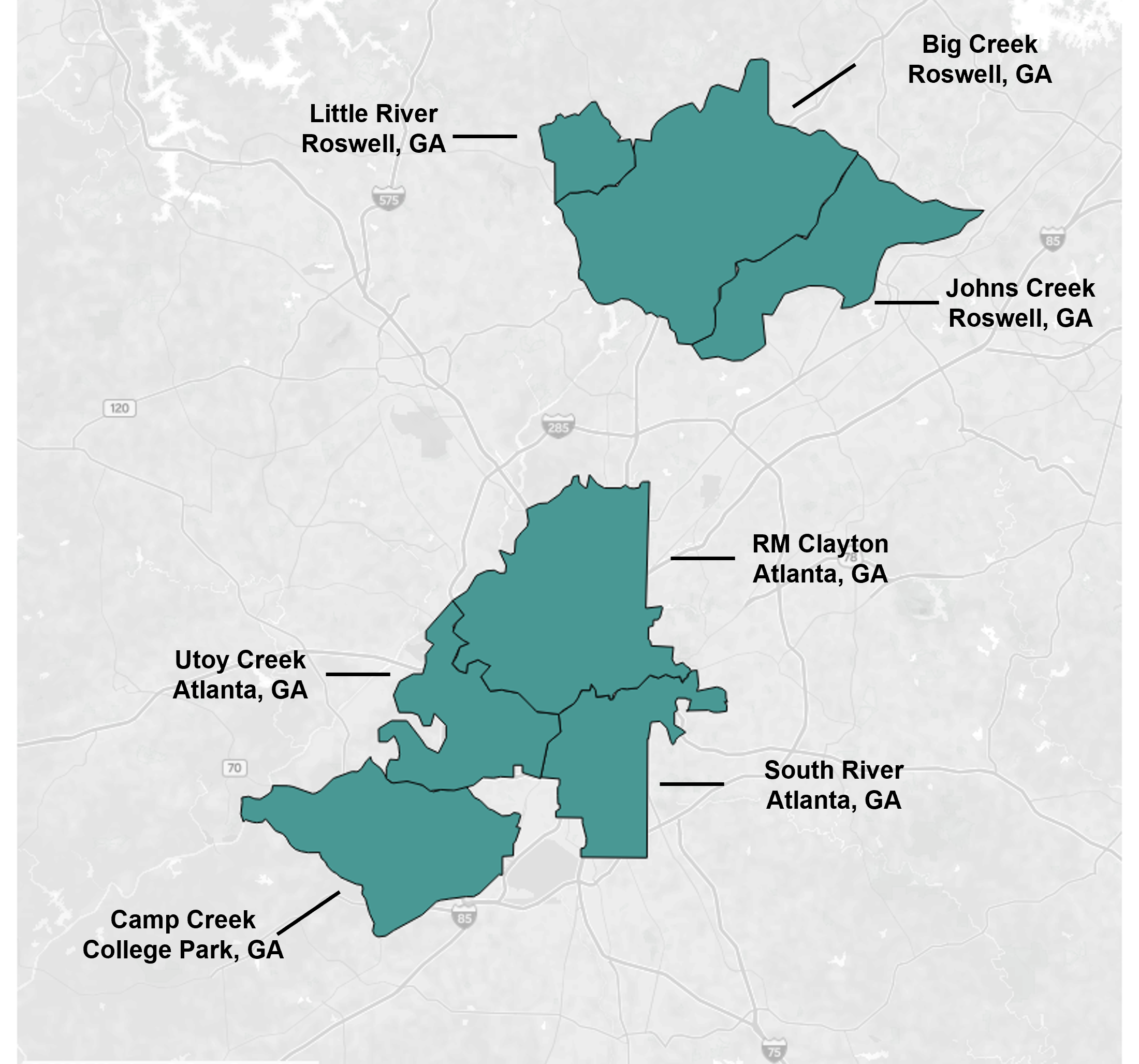
Georgia WWSCAN biweekly update 8/4/2023
Welcome to the bi-weekly update for WWSCAN partners in Georgia! The samples provided up through 8/2/23 have been processed in the lab and data are on the site at data.wastewaterscan.org.
If you notice any bugs on the site or have any comments about it, please continue to send your feedback via email Amanda Bidwell at albidwel@stanford.edu. We thank you for your partnership!
COVID-19
SARS-CoV-2 and Variants
SARS-CoV-2 N gene RNA concentrations are slightly increasing and range between 40,000-250,000 copies per gram. Click on each chart to further interact with a chart on data.wastewaterscan.org. The dark gray line in the chart below is a population-weighted aggregated trend line for all 8 Georgia sites when the data is normalized by PMMoV. Additional information on the methodology behind this trend line is available here in the "How are aggregated trendlines determined?" section. More details can be found under the "How are national levels determined?" section here. The aggregated line below shows that SARS-CoV-2 N gene RNA concentrations are currently within the middle third level of all concentrations measure in the last year.

For most targets, we are also including a trend analysis of their concentrations in wastewater solids (normalized by PMMoV) based on the most recent estimates compared to the recent past. In order to calculate a trend, we require 3 points over the last 21 days. We test for linear trends between log10-transformed concentration (target/PMMoV) and time; the trend must be classified as statistically significant (p<0.1). To read more about our methods for trend analysis click the link here. Analysis of SARS-CoV-2 concentrations across the 8 Georgia sites shows there is no trend at each site except for a significant upward trend seen at Little River.
Early in 2023, WWSCAN started testing for XBB* and these results are available to view by selecting 'SARS-CoV-2 View by Variant' on data.wastewaterscan.org. For Georgia sites, data dates back to early February 2023. The ratio of the XBB* mutations/N across all Georgia plants is shown in the chart below. As the ratio reaches 100%, it suggests that all the SARS-CoV-2 genomes in wastewater have the XBB* mutations. These estimates have not changed much since our last update, and data indicate that the average ratio XBB* mutations/N is ~75% but has ranged from 40-100% recently. This suggests the majority of infections across the communities are caused by XBB*. The next chart below shows the ratio of the BA.5, BA.4, and BQ* mutation/N across all Georgia plants. It can be read the same way as the XBB* chart. The average ratio is close to 0%. Note that our XBB* assay will also detect XBB.1.16, XBB.1.9, and FD.2 (XBB.1.5.15.2*).


Other Respiratory Viruses
IAV, IBV, RSV, and HMPV
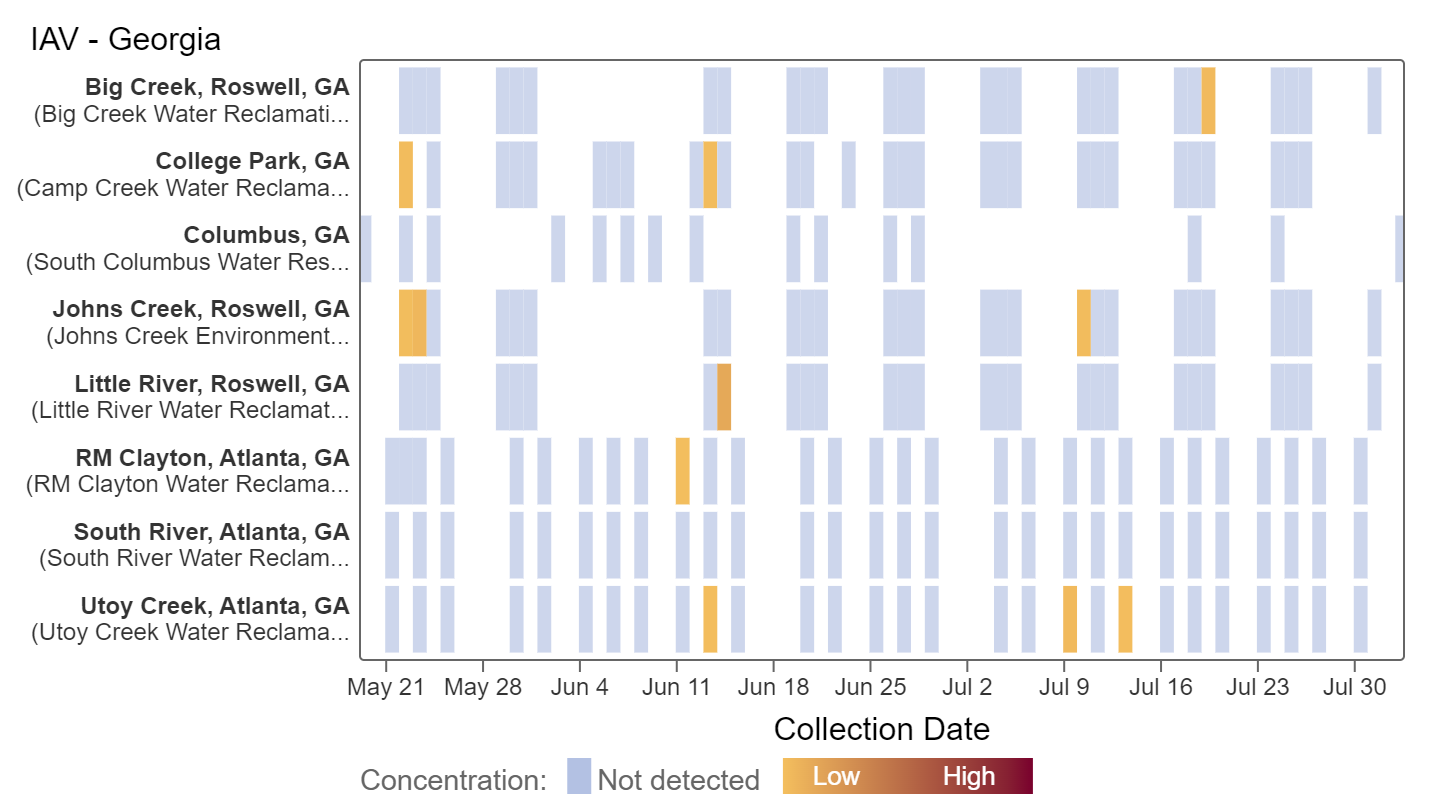
Influenza A (IAV) RNA has not been detected in samples collected in the last two weeks as seen in the heat map below. Analysis of IAV concentrations indicates there is no significant trend across all 8 Georgia sites. This can be interpreted to mean that over the last 21 days, concentrations of IAV RNA have generally not changed.
Influenza B (IBV) RNA also was not detected in the last two weeks and the analysis of IBV concentrations determined there is no significant trend across all 8 Georgia sites.

RSV RNA concentrations are low, which is shown below in this heat map. Big Creek and Utoy Creek have not detected RSV RNA in samples collected over the last two weeks. The analysis of RSV concentrations indicates there is no significant trend across all 8 Georgia sites.

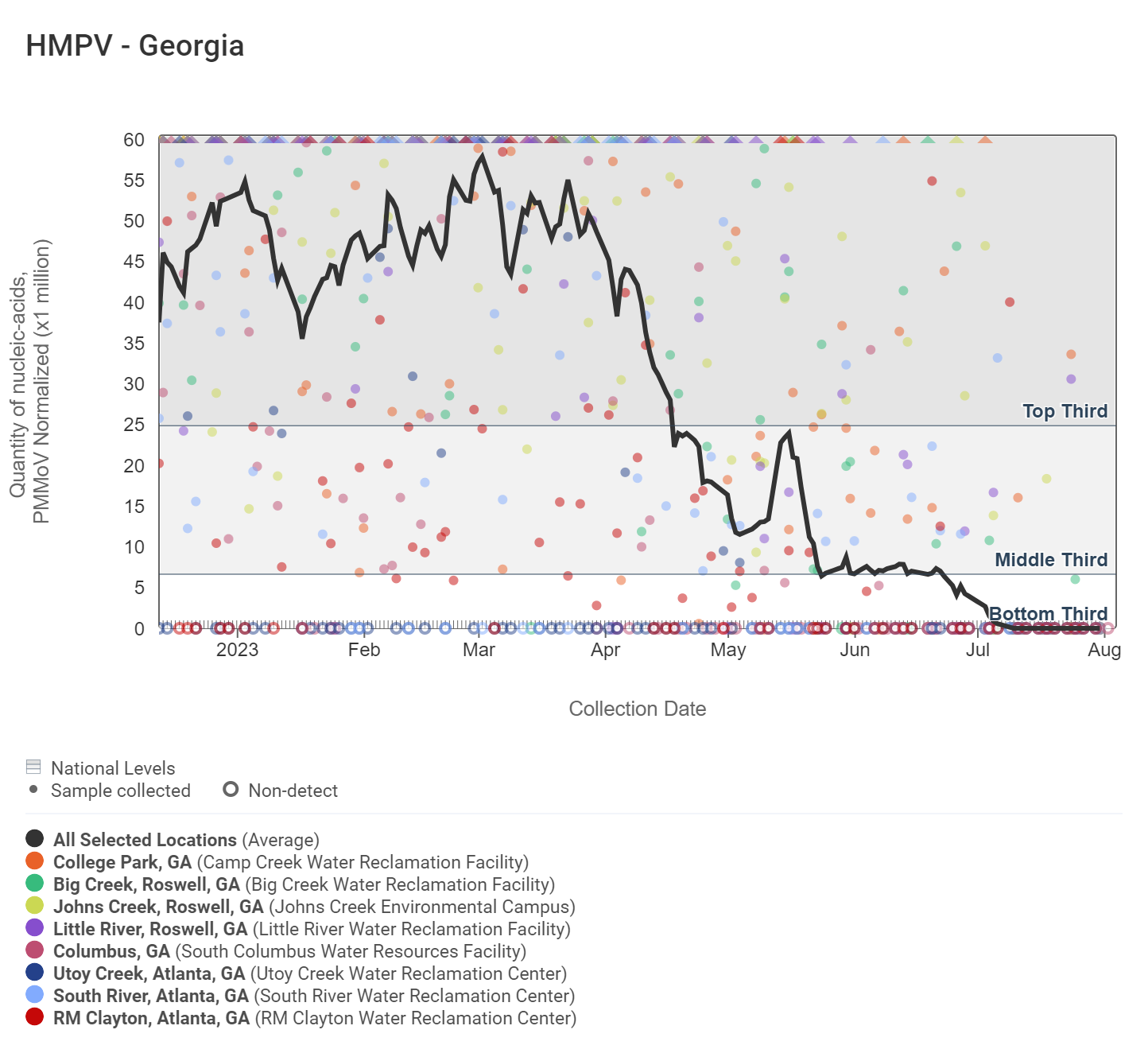
HMPV (human metapneumovirus) RNA concentrations have decreased and are increasingly non-detect and in the bottom third level according to the chart below. Big Creek, College Park, and Little River are the only sites that have detected HMPV within the last two weeks. The analysis of HMPV concentrations showed there is no significant trend across all Georgia sites.
Norovirus
Assay detects human norovirus GII
Norovirus GII (HuNoV GII) RNA concentrations range between 600,000 and 6,000,000 copies/g. An analysis of HuNoV GII concentrations showed there is no significant trend across Georgia sites. This can be interpreted to mean that over the last 21 days, concentrations of HuNoV GII RNA have generally not changed at most Georgia sites.

Mpox
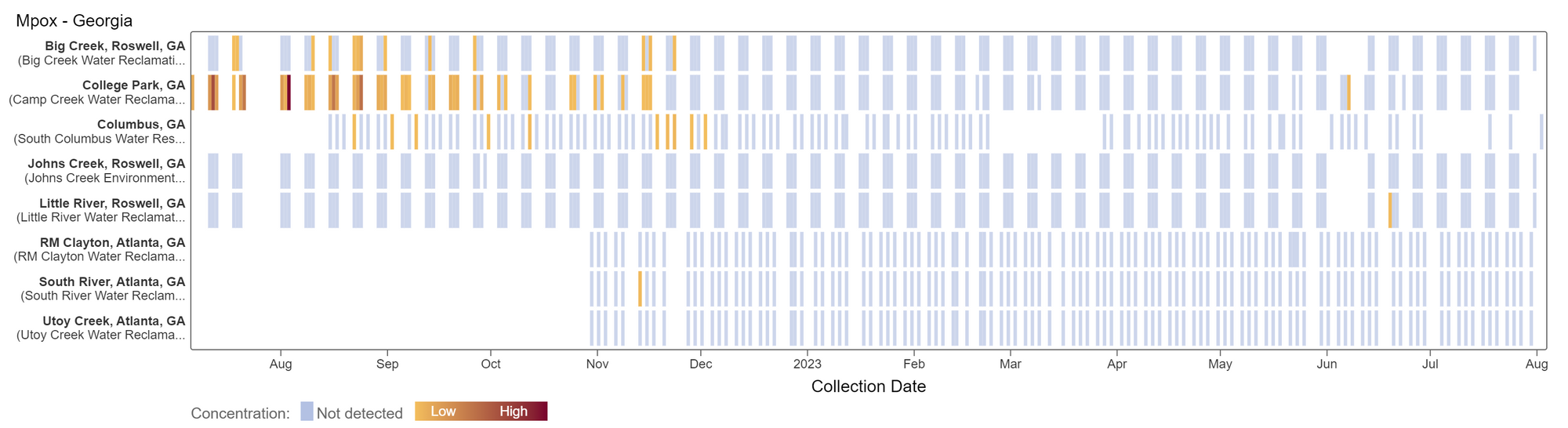
Results are non-detect for mpox for the past 21 days at Georgia plants. This heat map shows data since July 2022. Sites are labeled in the rows and each date a sample was collected as a column. The color blue means the sample was non-detect for mpox DNA and the colors get darker with higher concentrations.
Related News
The next stakeholder meeting will take place Friday, August 11th @ 12 PM EST. We hope to see you there!
