SCAN Weekly Update - 9/13/23
This blog post describes data collected at 8 wastewater treatment plants in the Greater Bay Area of San Francisco, CA, including Sacramento, that are partners in the SCAN project which began in late 2020.
There are three recent papers related to this work that you might be interested in:
L. Roldan-Hernandez and A. B. Boehm. 2023. Adsorption of Respiratory Syncytial Virus, Rhinovirus, SARS-CoV-2, and F+ Bacteriophage MS2 RNA onto Wastewater Solids from Raw Wastewater. Environmental Science & Technology. Link.
A. B. Boehm, M. K. Wolfe, B. J. White, B. Hughes, D. Duong, A. Bidwell. 2023. More than a Tripledemic: Influenza A Virus, Respiratory Syncytial Virus, SARS-CoV-2, and Human Metapneumovirus in Wastewater during Winter 2022 - 2023. Environmental Science & Technology Letters, 10, 8, 622-627. Link.
A. B. Boehm, M. K. Wolfe, B. J. White, B. Hughes, D. Duong, N. Banaei, A. Bidwell. Human norovirus (HuNoV) GII RNA in wastewater solids at 145 United States wastewater treatment plants: Comparison to positivity rates of clinical specimens and modeled estimates of HuNoV GII shedders. Journal of Exposure Science and Environmental Epidemiology. Link.

Map of 8 SCAN sites located in the Greater San Francisco Bay Area, CA
All samples picked up by the couriers as of 9/11/23 have been processed and their data are on the site: data.wastewaterscan.org. Please email Amanda Bidwell at albidwel@stanford.edu if you identify any bugs on the site.
Please note that we sunset measurement of influenza B virus (IBV) to make room for new targets. We began testing for enterovirus D68, rotavirus, human adenovirus group F, Candida auris, and hepatitis A in early July, and for parainfluenza on 9/5. We do not yet have enough data to provide information about trends. Over the past week, we have had mostly non-detects for enterovirus D68 across SCAN plants. In contrast, parainfluenza, human adenovirus group F, and rotavirus have been detected in most samples from SCAN plants. C. auris and hepatitis A have been detected sporadically at most SCAN plants. At San Jose specifically, hepatitis A has been detected in most samples.
COVID-19
SARS-CoV-2 and Variants
SARS-CoV-2 N gene concentrations are between 10,000 and 100,000 copies/g. Below is a chart from all the SCAN plants that highlights the centered, 5-d trimmed average values (green line) and the raw data (green points) at Oceanside SF. The black vertical line denotes 3 weeks prior to today.

We ran a 3-week look period trend analysis on the plants for the N gene target using methods outlined in this peer-reviewed paper by our team (link to paper). The 3-week look period trend analysis on SARS-CoV-2 at these plants indicates a significant downward trend at Oceanside and no trend at all other individual SCAN plants.
Here are all the data from all the SCAN plants for the last two years. You can access the graph here. The population weighted average across the SCAN plants is shown in black. Oceanside has the highest concentrations represented by the line above the rest.
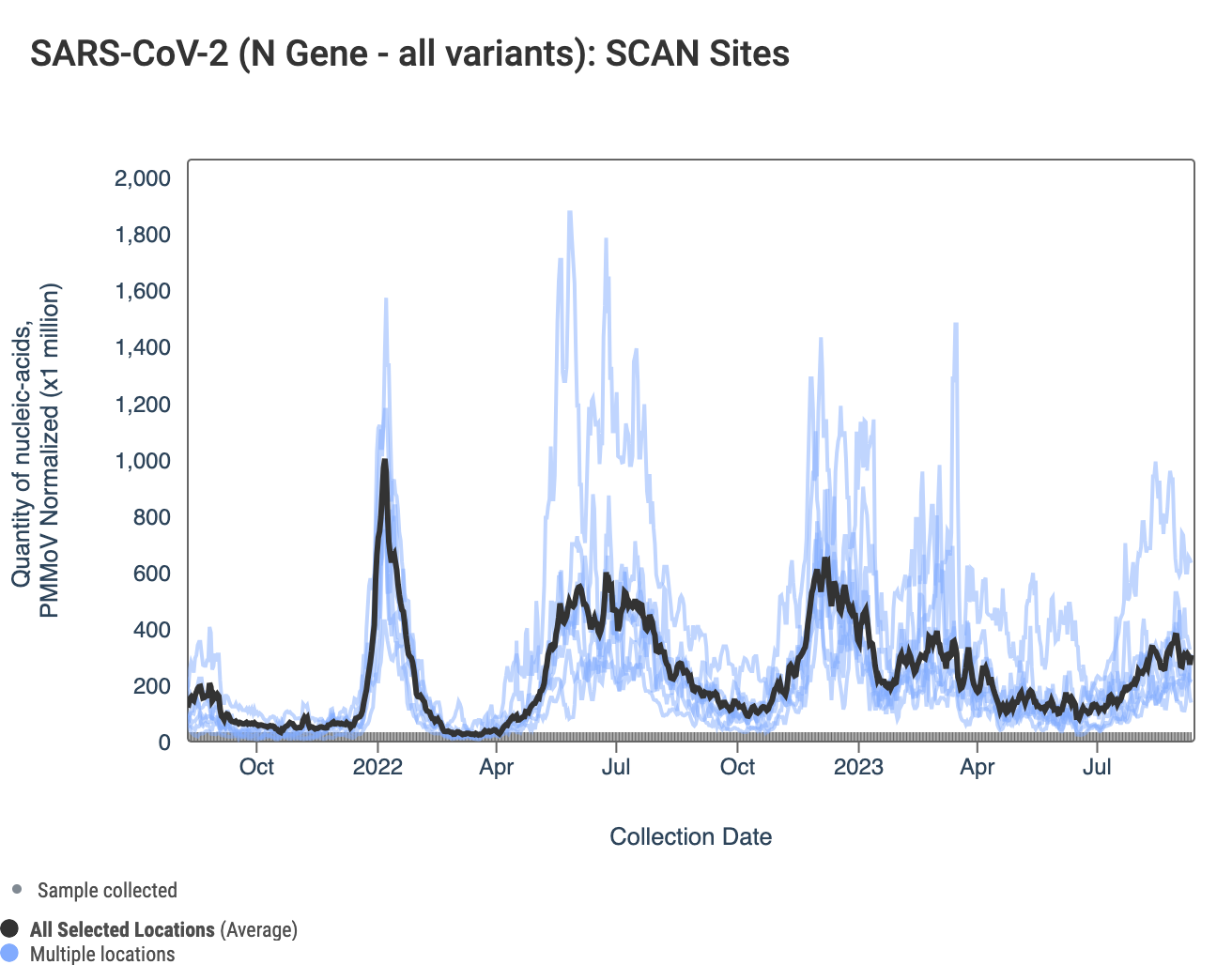
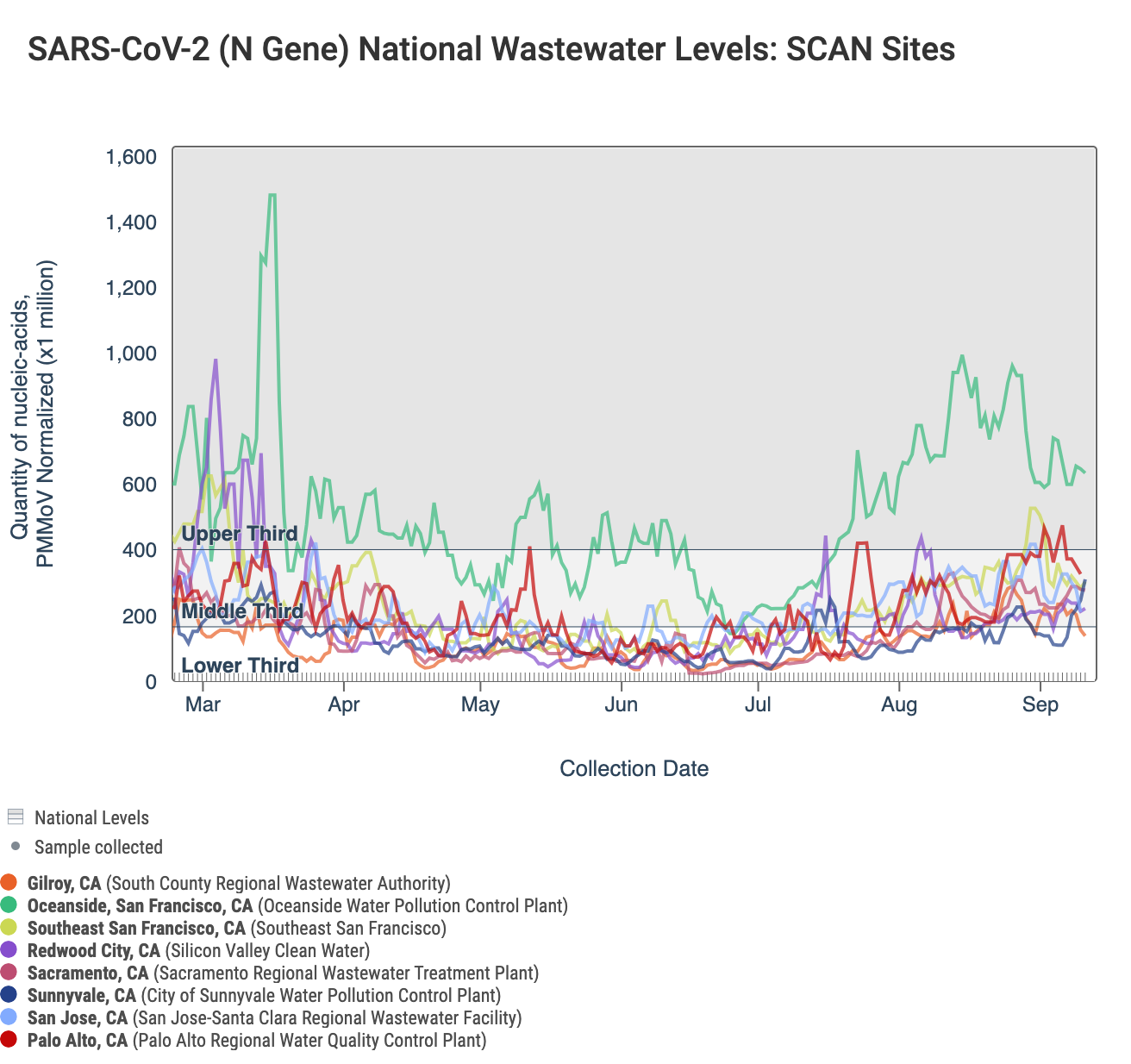
Below is a chart showing the National Levels benchmarks superimposed on the SARS-CoV-2 N gene data from the SCAN sites over the last 6 weeks. Currently SCAN sites SARS-CoV-2 N gene concentrations at Oceanside are in the top third level, Gilroy is in the lower third level, and all other locations are in the middle third level. You can access the graph here.
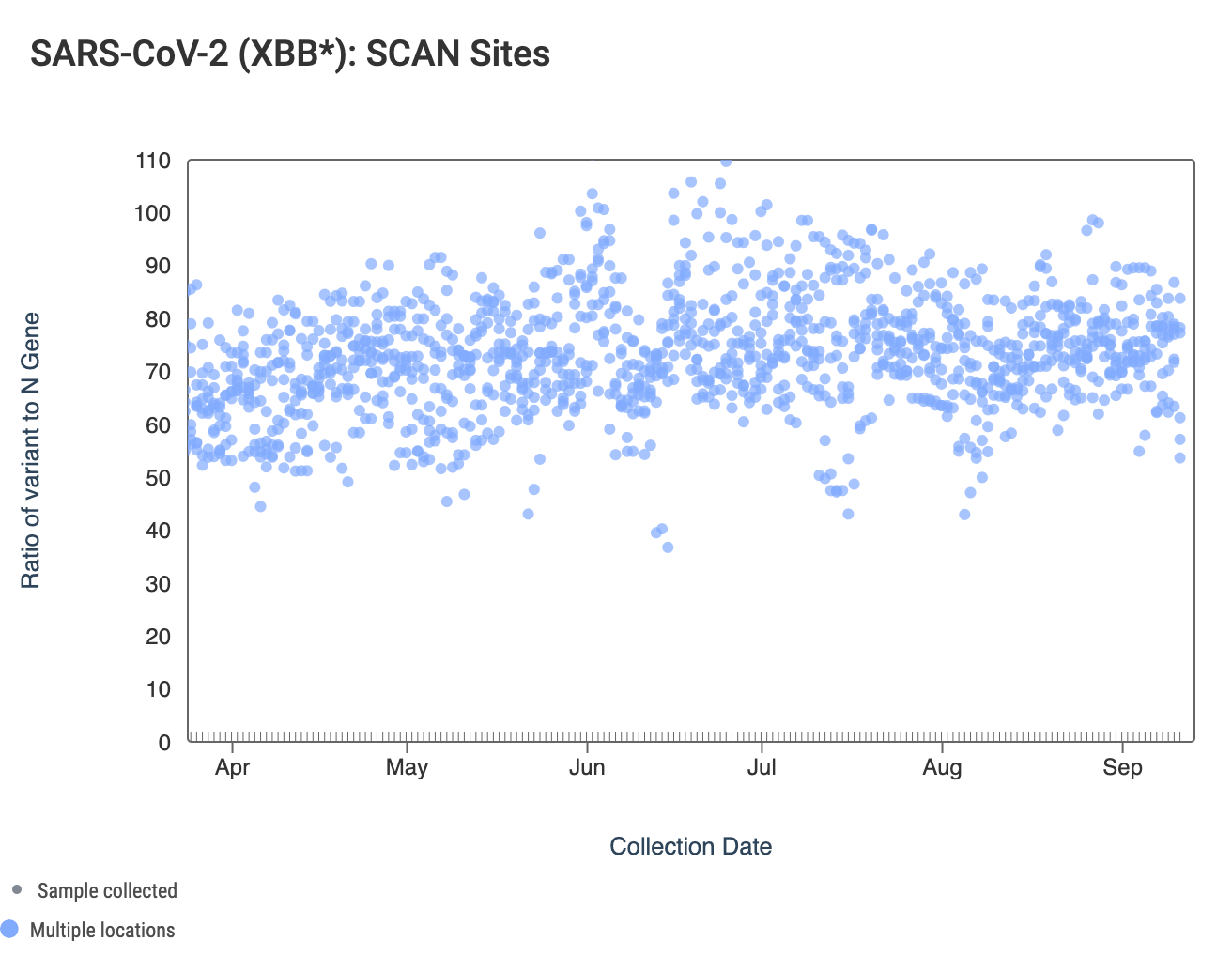
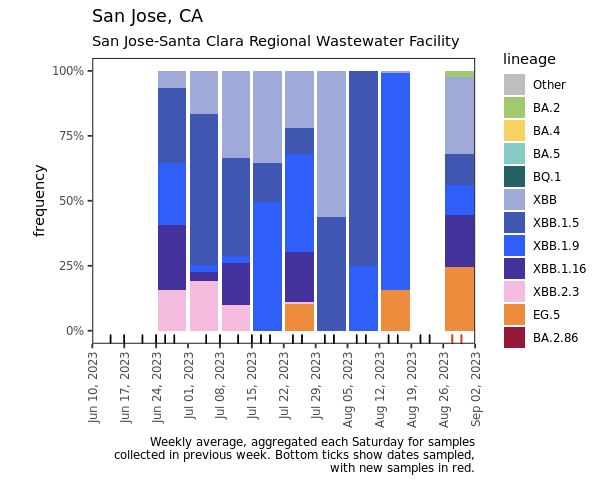
The ratio of the XBB* mutations/N across SCAN plants is shown in the chart below. This chart comes from data.wastewaterscan.org (link here to the chart). As the ratio reaches 100%, it suggests that all the SARS-CoV-2 genomes in wastewater have the XBB* mutations. These data indicate that the regional average ratio XBB* mutations/N is ~50-90%. These data suggest that the majority of infections across the SCAN communities are caused by XBB*. Note that these set of mutations are also present in EG.5 sublineage, but they are not present in BA.2.86.
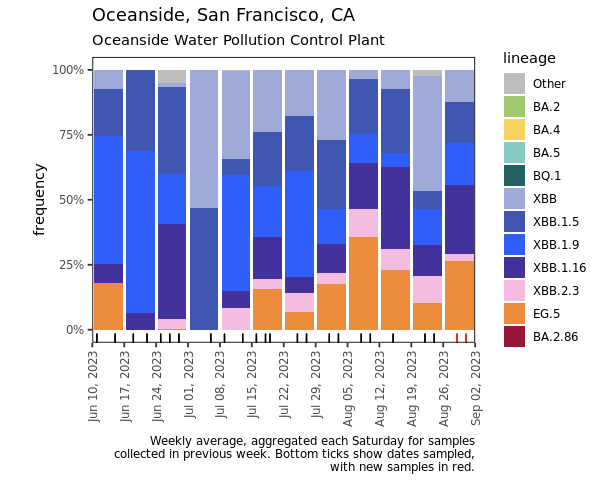
Below are plots from Oceanside, Sacramento, and San Jose showing the relative proportions of different variants inferred from sequencing the entire genome of SARS-CoV-2. Samples with low coverage are not being displayed in the plots (this explains some empty columns for Sacramento and San Jose). You can see that that all 3 plats had around 25% of the sublineage EG.5 (in orange) in the most recent sample. EG.5 is an Omicron sublineage that has emerged in other parts of the world and has been causing infections in the US. You can read more about it here. Note that the sequencing data are always from samples taken between 1- 2 weeks ago. We have not detected BA.2.86 in any samples yet, but it should be easy to detect when it is present because it has some characteristic deletions and a large insertion. BA.2.86 was detected in a wastewater sample somewhere, as describe in this press release from CDC.

Other Respiratory Targets
Influenza A & B, RSV, and HMPV
Influenza A (IAV) RNA concentrations are on average higher than they were at this time last year. In the chart below, the population weighted average line across all the SCAN plants is shown in black, and you can see a recent small uptick. The link to the chart below is here if you would like to interact with it.
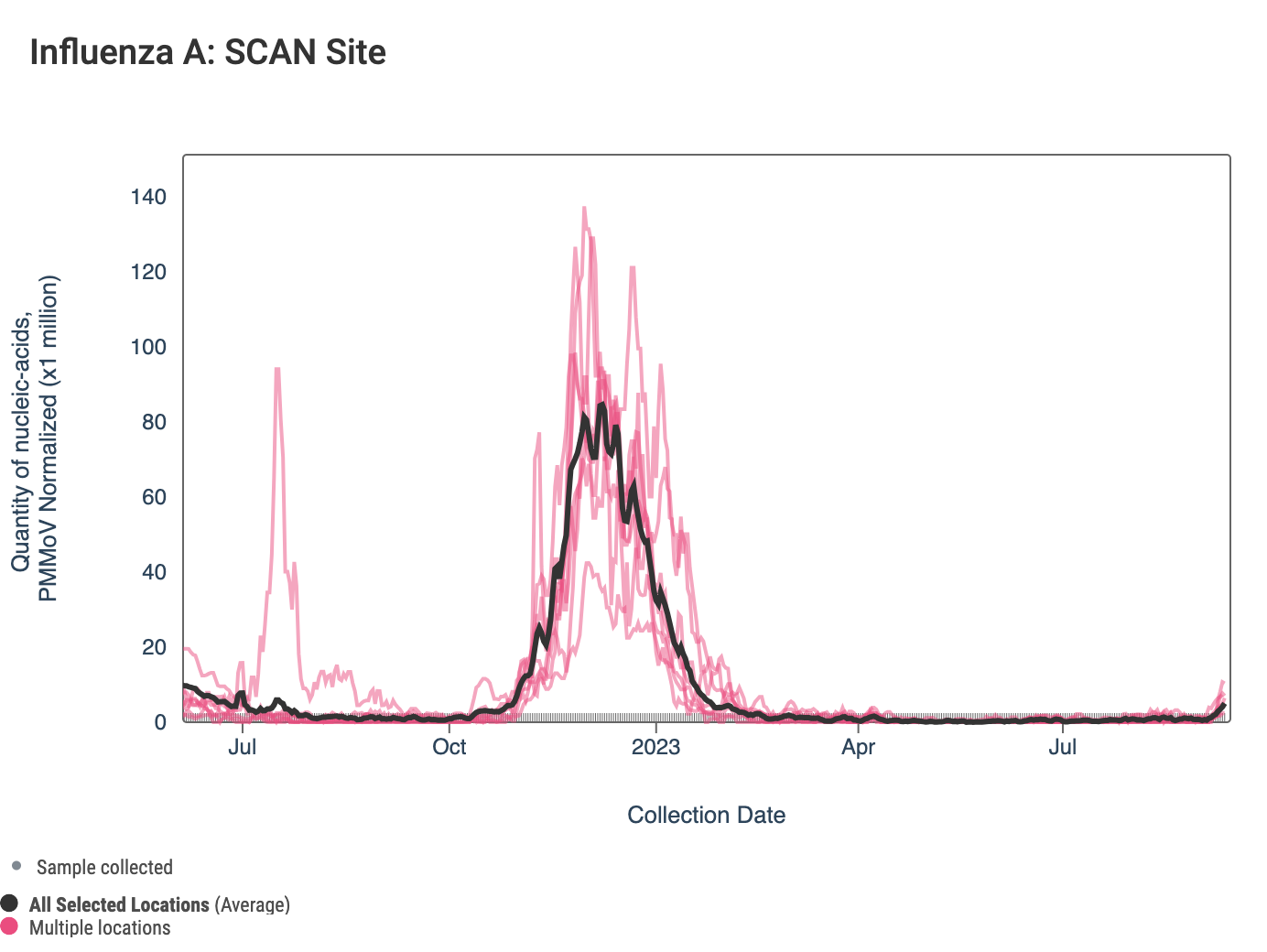
The 3-week look period trend analysis on IAV RNA at these plants indicates no trend at all the SCAN plants.
RSV RNA concentrations are shown from all the SCAN sites in the chart below together since last winter and can be access here. The black line is the population weighted average. On average, concentrations are higher than they were this time last year.
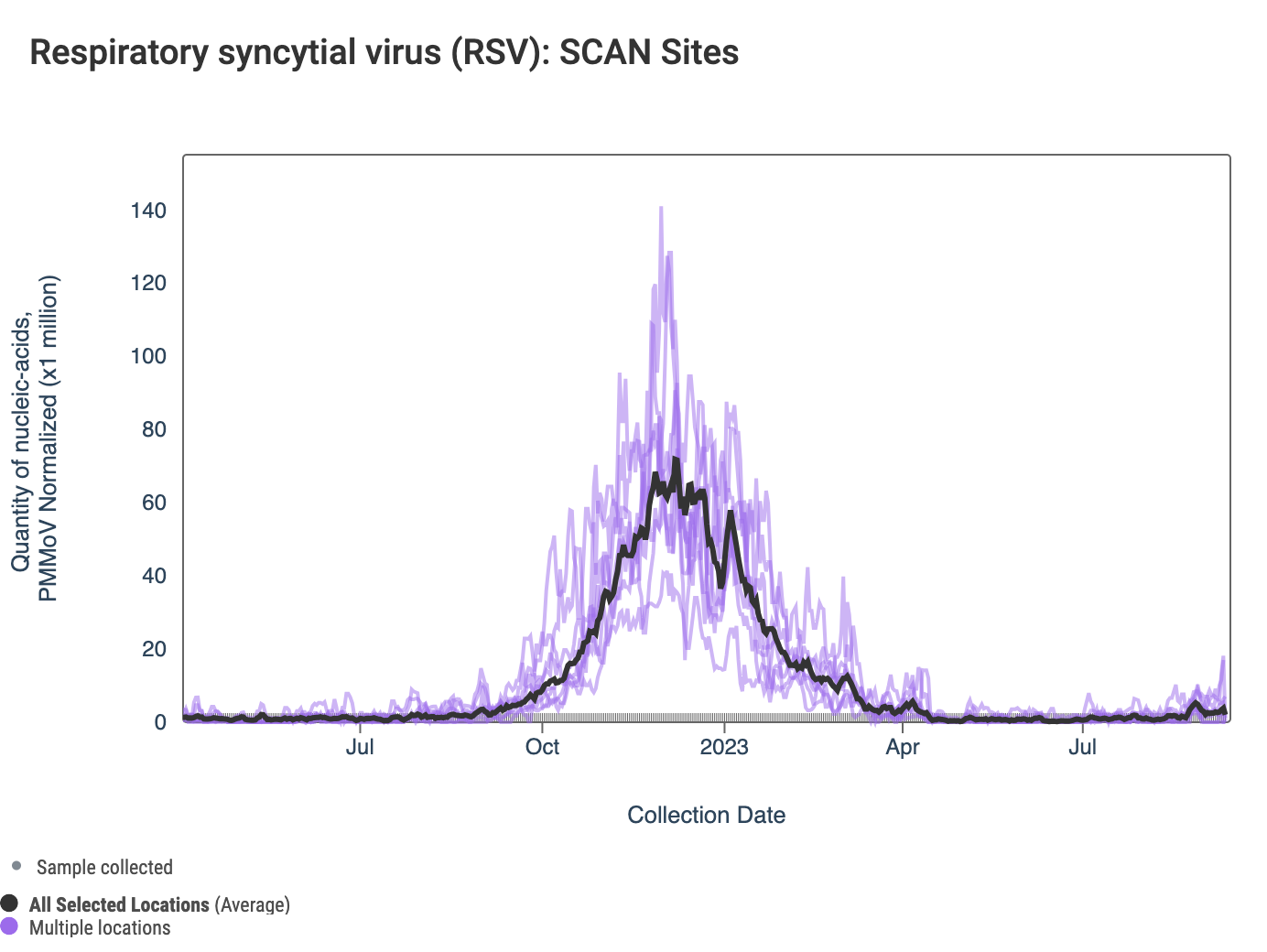
The 3-week look period trend analysis on RSV RNA shows no significant trends at any SCAN plant.
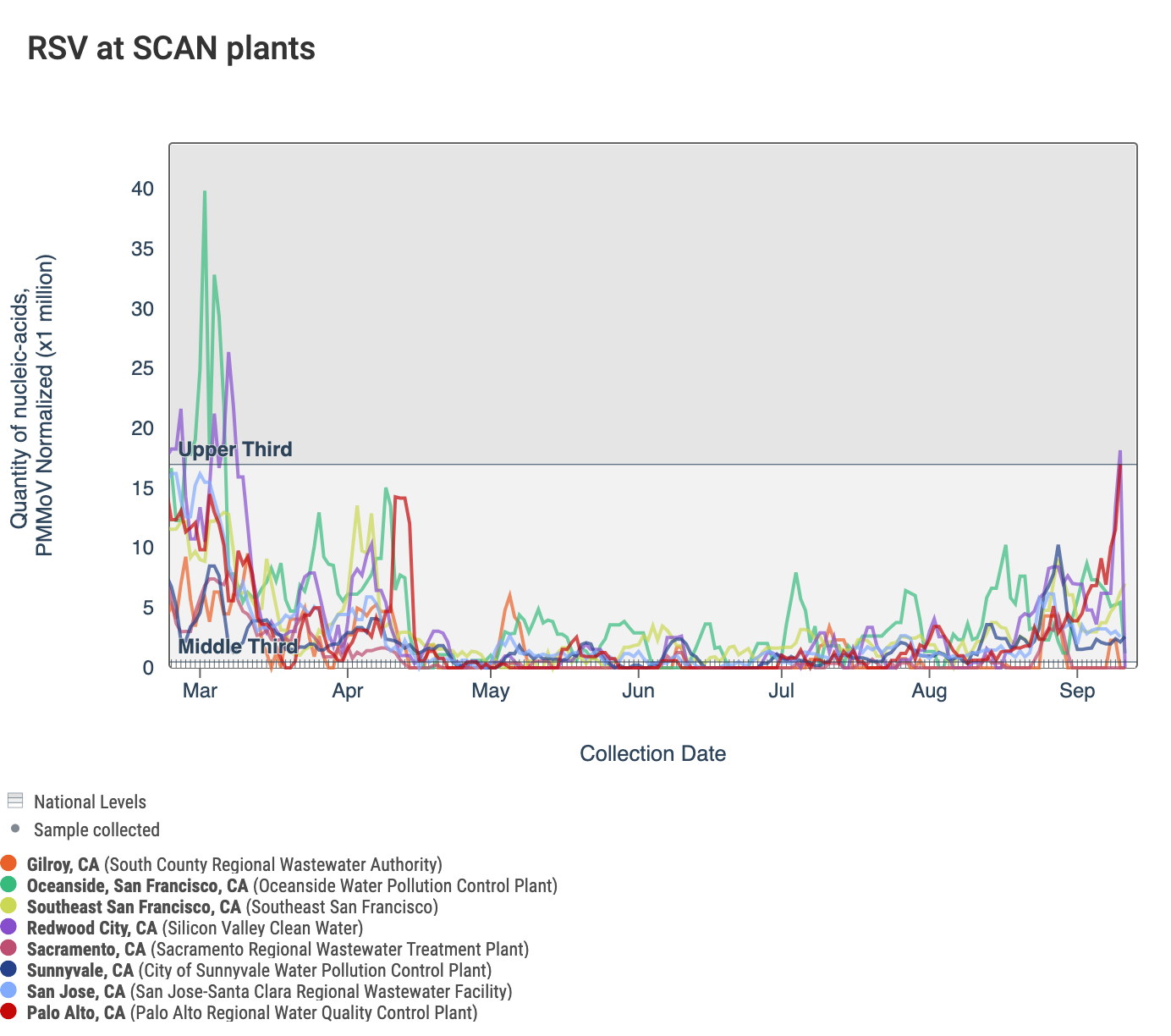
Below is a chart showing the National Levels benchmarks superimposed on the RSV data from the SCAN sites over the last 6 months (you can access this chart here). Currently, the RSV concentration is approaching the upper third for Palo Alto and is in the middle third for Oceanside, San Jose, Southeast San Francisco, and Sunnyvale. The RSV concentration at Gilroy, Sacramento, and Redwood City is currently in the bottom third, but Redwood City has had concentrations at all three levels during the last week.
HMPV (human metapneumovirus) RNA concentrations are low at most of plants in SCAN. Below are charts showing data from all the SCAN plants together (link here to this chart if you want to interact with it).
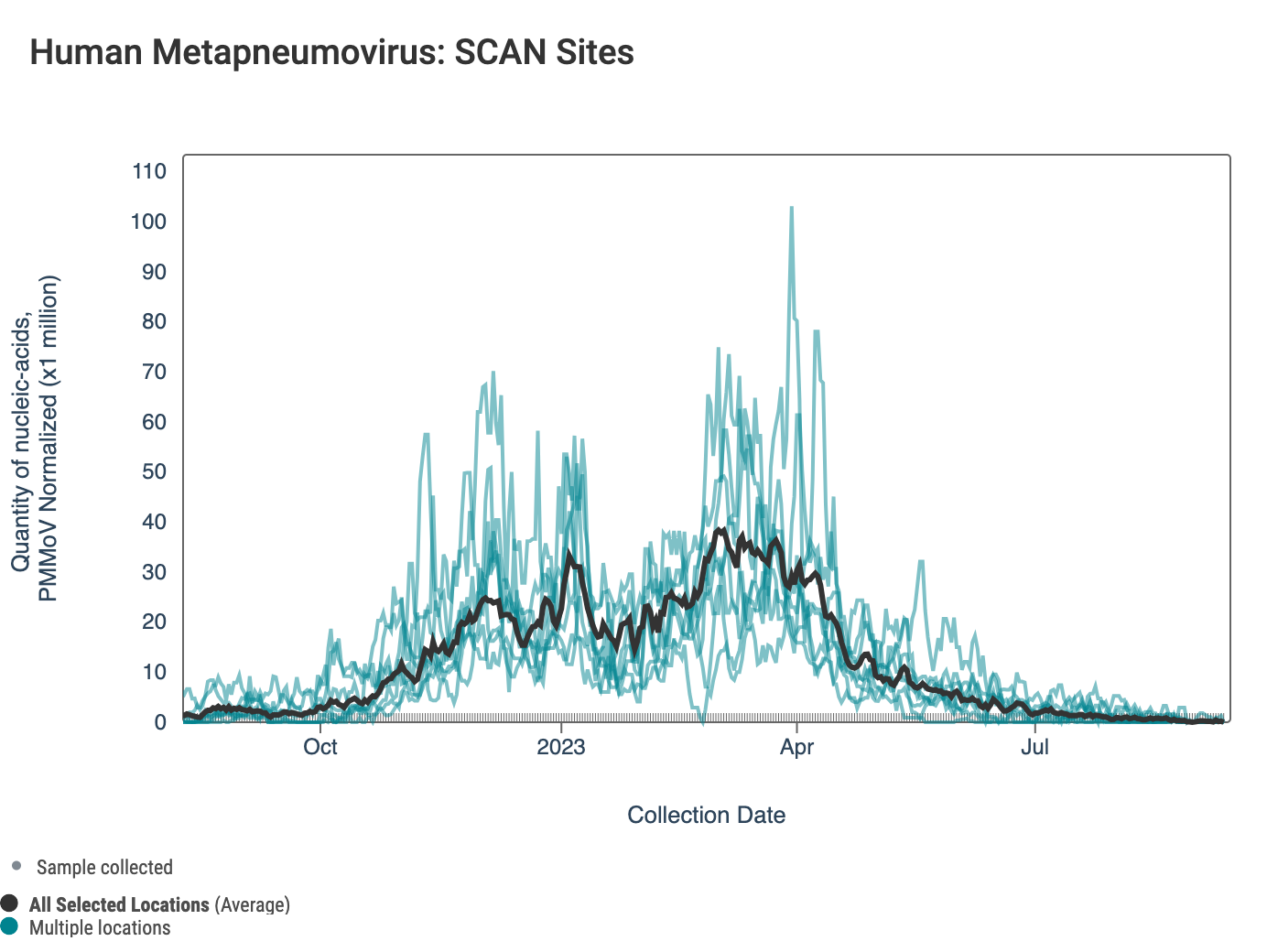
The 3-week look period trend analysis on HMPV RNA shows no significant trends at any SCAN plant.
Below is a chart showing the National Levels benchmarks superimposed on the HMPV data from the SCAN sites over the last 6 months (you can access this chart here). Currently, the HMPV concentration is in the lower third level for al SCAN plants.

Gastrointestinal Targets
Norovirus GII
Norovirus GII RNA concentrations are shown in the chart below; all the SCAN plants are shown together. The population weighted average line is shown in black. You can interact with the chart of all the plants at this link.
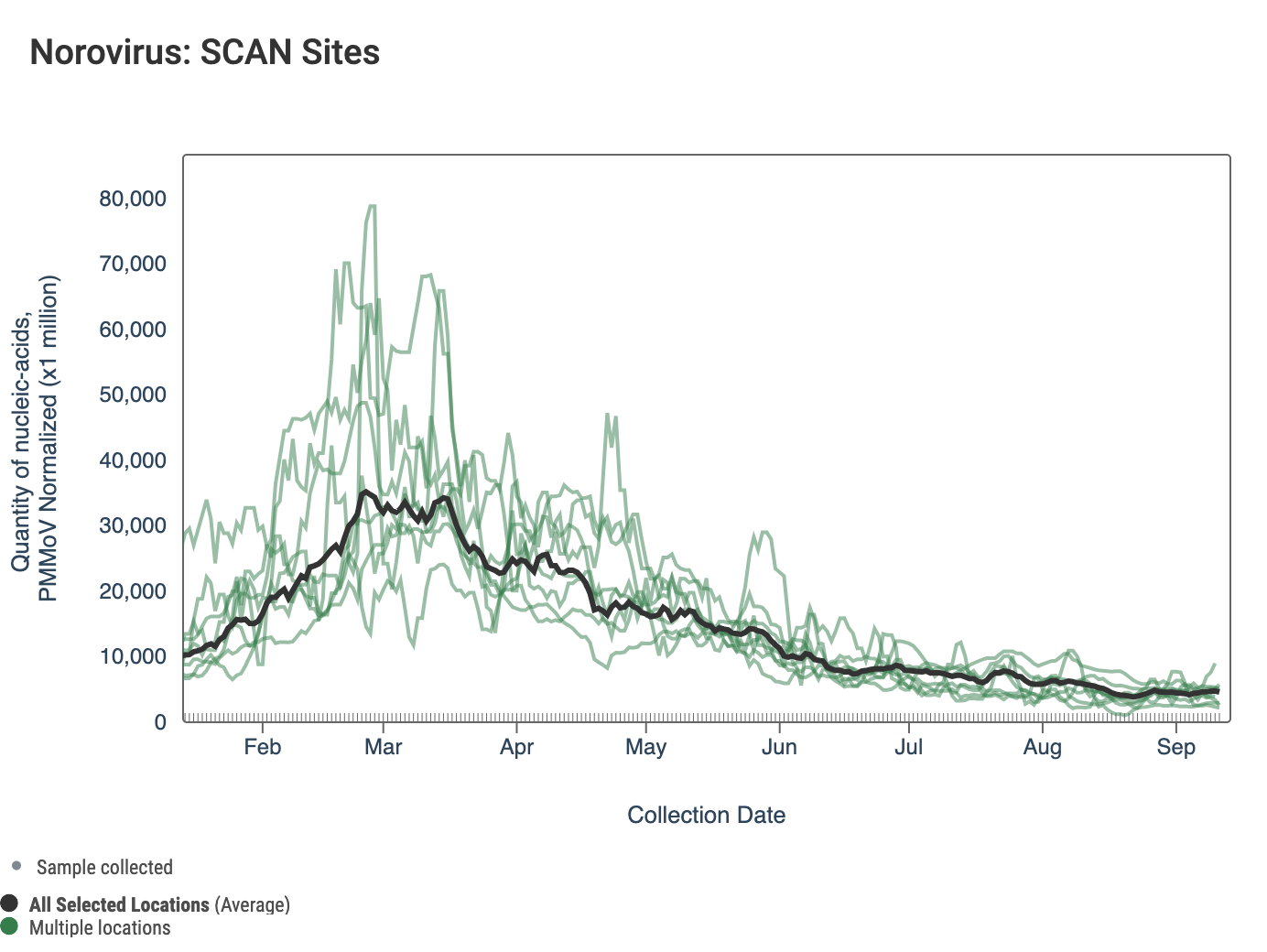
The 3-week look period trend analysis on HuNoV GII RNA at SCAN plants indicates a significant downward trend for Oceanside, Sunnyvale, and Southeast San Francisco and no trend for all the other SCAN plants.
Below is a chart showing the National Levels benchmarks superimposed on the HuNoV GII data from the SCAN sites since the beginning of 2023. (You can access the chart here.) Currently all SCAN sites HuNoV GII concentrations in the lower third level.

Other Pathogens of Concern
Mpox
We've observed non-detects in the last 14 days for MPXV DNA at the SCAN plants. The heat map show all the SCAN sites as a row, and each date as a column. The color blue means the sample was non-detect for MPXV DNA and the colors get darker with higher concentrations. White indicates no sample was collected. The charts shows all the data we have collected, almost a year's worth of data. You can access the chart here.

